Pim-1 Antibody - #AF0844
| Product: | Pim-1 Antibody |
| Catalog: | AF0844 |
| Description: | Rabbit polyclonal antibody to Pim-1 |
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 50kDa; 36kD(Calculated). |
| Uniprot: | P11309 |
| RRID: | AB_2834110 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0844, RRID:AB_2834110.
Fold/Unfold
Oncogene PIM 1; Oncogene PIM1; PIM 1; pim 1 kinase 44 kDa isoform; Pim 1 kinase; pim 1 oncogene (proviral integration site 1); Pim 1 oncogene; PIM; PIM1; pim1 kinase 44 kDa isoform; PIM1_HUMAN; Pim2; PIM3; Proto oncogene serine/threonine protein kinase Pim 1; Proto-oncogene serine/threonine-protein kinase Pim-1; Proviral integration site 1; Proviral integration site 2;
Immunogens
Expressed primarily in cells of the hematopoietic and germline lineages. Isoform 1 and isoform 2 are both expressed in prostate cancer cell lines.
- P11309 PIM1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MLLSKINSLAHLRAAPCNDLHATKLAPGKEKEPLESQYQVGPLLGSGGFGSVYSGIRVSDNLPVAIKHVEKDRISDWGELPNGTRVPMEVVLLKKVSSGFSGVIRLLDWFERPDSFVLILERPEPVQDLFDFITERGALQEELARSFFWQVLEAVRHCHNCGVLHRDIKDENILIDLNRGELKLIDFGSGALLKDTVYTDFDGTRVYSPPEWIRYHRYHGRSAAVWSLGILLYDMVCGDIPFEHDEEIIRGQVFFRQRVSSECQHLIRWCLALRPSDRPTFEEIQNHPWMQDVLLPQETAEIHLHSLSPGPSK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P11309 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K5 | Ubiquitination | Uniprot | |
| S8 | Phosphorylation | Uniprot | |
| T23 | Phosphorylation | Uniprot | |
| K24 | Ubiquitination | Uniprot | |
| K31 | Ubiquitination | Uniprot | |
| S46 | Phosphorylation | Uniprot | |
| K67 | Ubiquitination | Uniprot | |
| K94 | Ubiquitination | Uniprot | |
| S98 | Phosphorylation | Uniprot | |
| K169 | Ubiquitination | Uniprot | |
| K183 | Ubiquitination | Uniprot | |
| S189 | Phosphorylation | P11309 (PIM1) | Uniprot |
| K194 | Ubiquitination | Uniprot | |
| S261 | Phosphorylation | Uniprot | |
| S308 | Phosphorylation | Uniprot |
PTMs - P11309 As Enzyme
| Substrate | Site | Source |
|---|---|---|
| O14757 (CHEK1) | S280 | Uniprot |
| O43524 (FOXO3) | T32 | Uniprot |
| O43524 (FOXO3) | S253 | Uniprot |
| P01106 (MYC) | S329 | Uniprot |
| P10275 (AR) | S215 | Uniprot |
| P10275 (AR) | T851 | Uniprot |
| P11309 (PIM1) | S189 | Uniprot |
| P27448-3 (MARK3) | T90 | Uniprot |
| P27448 (MARK3) | T95 | Uniprot |
| P27448-3 (MARK3) | S96 | Uniprot |
| P30304-1 (CDC25A) | S116 | Uniprot |
| P36888 (FLT3) | Y591 | Uniprot |
| P38936 (CDKN1A) | T145 | Uniprot |
| P38936 (CDKN1A) | S146 | Uniprot |
| P46527 (CDKN1B) | T157 | Uniprot |
| P46527 (CDKN1B) | T198 | Uniprot |
| P61073 (CXCR4) | S339 | Uniprot |
| P68431 (HIST1H3J) | S11 | Uniprot |
| Q00987 (MDM2) | S166 | Uniprot |
| Q00987 (MDM2) | S186 | Uniprot |
| Q01860 (POU5F1) | S288 | Uniprot |
| Q01860 (POU5F1) | S289 | Uniprot |
| Q04206 (RELA) | S276 | Uniprot |
| Q12778 (FOXO1) | T24 | Uniprot |
| Q12778 (FOXO1) | S256 | Uniprot |
| Q12778 (FOXO1) | S319 | Uniprot |
| Q13309 (SKP2) | S64 | Uniprot |
| Q13309 (SKP2) | S72 | Uniprot |
| Q13309 (SKP2) | T417 | Uniprot |
| Q13761 (RUNX3) | S149 | Uniprot |
| Q13761 (RUNX3) | T151 | Uniprot |
| Q13761 (RUNX3) | T153 | Uniprot |
| Q13761 (RUNX3) | T155 | Uniprot |
| Q8TA86 (RP9) | S212 | Uniprot |
| Q8TA86 (RP9) | S214 | Uniprot |
| Q92934 (BAD) | S75 | Uniprot |
| Q92934 (BAD) | S99 | Uniprot |
| Q92934 (BAD) | S118 | Uniprot |
| Q96B36 (AKT1S1) | T246 | Uniprot |
| Q99683 (MAP3K5) | S83 | Uniprot |
| Q99801 (NKX3-1) | T89 | Uniprot |
| Q99801 (NKX3-1) | S185 | Uniprot |
| Q99801 (NKX3-1) | S186 | Uniprot |
| Q99801 (NKX3-1) | S195 | Uniprot |
| Q99801 (NKX3-1) | S196 | Uniprot |
| Q9BZS1 (FOXP3) | S422 | Uniprot |
| Q9UNQ0 (ABCG2) | T362 | Uniprot |
Research Backgrounds
Proto-oncogene with serine/threonine kinase activity involved in cell survival and cell proliferation and thus providing a selective advantage in tumorigenesis. Exerts its oncogenic activity through: the regulation of MYC transcriptional activity, the regulation of cell cycle progression and by phosphorylation and inhibition of proapoptotic proteins (BAD, MAP3K5, FOXO3). Phosphorylation of MYC leads to an increase of MYC protein stability and thereby an increase of transcriptional activity. The stabilization of MYC exerted by PIM1 might explain partly the strong synergism between these two oncogenes in tumorigenesis. Mediates survival signaling through phosphorylation of BAD, which induces release of the anti-apoptotic protein Bcl-X(L)/BCL2L1. Phosphorylation of MAP3K5, an other proapoptotic protein, by PIM1, significantly decreases MAP3K5 kinase activity and inhibits MAP3K5-mediated phosphorylation of JNK and JNK/p38MAPK subsequently reducing caspase-3 activation and cell apoptosis. Stimulates cell cycle progression at the G1-S and G2-M transitions by phosphorylation of CDC25A and CDC25C. Phosphorylation of CDKN1A, a regulator of cell cycle progression at G1, results in the relocation of CDKN1A to the cytoplasm and enhanced CDKN1A protein stability. Promotes cell cycle progression and tumorigenesis by down-regulating expression of a regulator of cell cycle progression, CDKN1B, at both transcriptional and post-translational levels. Phosphorylation of CDKN1B, induces 14-3-3 proteins binding, nuclear export and proteasome-dependent degradation. May affect the structure or silencing of chromatin by phosphorylating HP1 gamma/CBX3. Acts also as a regulator of homing and migration of bone marrow cells involving functional interaction with the CXCL12-CXCR4 signaling axis. Also phosphorylates and activates the ATP-binding cassette transporter ABCG2, allowing resistance to drugs through their excretion from cells.
Autophosphorylated on both serine/threonine and tyrosine residues. Phosphorylated. Interaction with PPP2CA promotes dephosphorylation.
Ubiquitinated, leading to proteasomal degradation.
Cytoplasm. Nucleus.
Cell membrane.
Expressed primarily in cells of the hematopoietic and germline lineages. Isoform 1 and isoform 2 are both expressed in prostate cancer cell lines.
Isoform 1 is isolated as a monomer whereas isoform 2 complexes with other proteins (By similarity). Binds to RP9 (By similarity). Isoform 2, but not isoform 1, binds BMX. Isoform 1 interacts with CDKN1B and FOXO3. Interacts with BAD (By similarity). Interacts with PPP2CA; this interaction promotes dephosphorylation of PIM1, ubiquitination and proteasomal degradation. Interacts with HSP90AA1, this interaction stabilizes PIM1 protein levels. Interacts (ubiquitinated form) with HSP70 and promotes its proteosomal degradation. Interacts with CDKN1A. Interacts with CDC25C. Interacts (via N-terminal 96 residues) with CDC25A (By similarity). Interacts with MAP3K5. Interacts with MYC (By similarity). Interacts with CBX3.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. PIM subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
References
Application: WB Species: Human Sample:
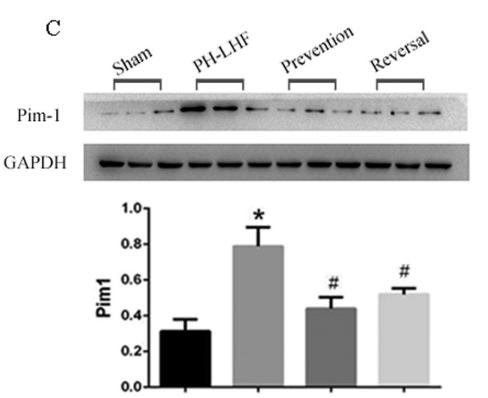
Application: WB Species: rat Sample: lung
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.