NMDAR2B Antibody - #AF6426
| Product: | NMDAR2B Antibody |
| Catalog: | AF6426 |
| Description: | Rabbit polyclonal antibody to NMDAR2B |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 180kDa; 166kD(Calculated). |
| Uniprot: | Q13224 |
| RRID: | AB_2835159 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6426, RRID:AB_2835159.
Fold/Unfold
AW490526; EIEE27; Glutamate [NMDA] receptor subunit epsilon 2; Glutamate [NMDA] receptor subunit epsilon-2; Glutamate Receptor Ionotropic N Methyl D Aspartate 2B; Glutamate Receptor Ionotropic N Methyl D Aspartate subunit 2B; Glutamate receptor ionotropic NMDA2B; Glutamate receptor subunit epsilon 2; Glutamate receptor, ionotropic, NMDA2B (epsilon 2); GRIN 2B; GRIN2B; hNR 3; hNR3; MGC142178; MGC142180; MRD6; N methyl D asparate receptor channel subunit epsilon 2; N methyl D aspartate receptor subtype 2B; N methyl D aspartate receptor subunit 2B; N methyl D aspartate receptor subunit 3; N-methyl D-aspartate receptor subtype 2B; N-methyl-D-aspartate receptor subunit 3; NMDA NR2B; NMDA R2B; NMDAR2B; NMDE2; NMDE2_HUMAN; NME2; NR2B; NR3;
Immunogens
Primarily found in the fronto-parieto-temporal cortex and hippocampus pyramidal cells, lower expression in the basal ganglia.
- Q13224 NMDE2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MKPRAECCSPKFWLVLAVLAVSGSRARSQKSPPSIGIAVILVGTSDEVAIKDAHEKDDFHHLSVVPRVELVAMNETDPKSIITRICDLMSDRKIQGVVFADDTDQEAIAQILDFISAQTLTPILGIHGGSSMIMADKDESSMFFQFGPSIEQQASVMLNIMEEYDWYIFSIVTTYFPGYQDFVNKIRSTIENSFVGWELEEVLLLDMSLDDGDSKIQNQLKKLQSPIILLYCTKEEATYIFEVANSVGLTGYGYTWIVPSLVAGDTDTVPAEFPTGLISVSYDEWDYGLPARVRDGIAIITTAASDMLSEHSFIPEPKSSCYNTHEKRIYQSNMLNRYLINVTFEGRNLSFSEDGYQMHPKLVIILLNKERKWERVGKWKDKSLQMKYYVWPRMCPETEEQEDDHLSIVTLEEAPFVIVESVDPLSGTCMRNTVPCQKRIVTENKTDEEPGYIKKCCKGFCIDILKKISKSVKFTYDLYLVTNGKHGKKINGTWNGMIGEVVMKRAYMAVGSLTINEERSEVVDFSVPFIETGISVMVSRSNGTVSPSAFLEPFSADVWVMMFVMLLIVSAVAVFVFEYFSPVGYNRCLADGREPGGPSFTIGKAIWLLWGLVFNNSVPVQNPKGTTSKIMVSVWAFFAVIFLASYTANLAAFMIQEEYVDQVSGLSDKKFQRPNDFSPPFRFGTVPNGSTERNIRNNYAEMHAYMGKFNQRGVDDALLSLKTGKLDAFIYDAAVLNYMAGRDEGCKLVTIGSGKVFASTGYGIAIQKDSGWKRQVDLAILQLFGDGEMEELEALWLTGICHNEKNEVMSSQLDIDNMAGVFYMLGAAMALSLITFICEHLFYWQFRHCFMGVCSGKPGMVFSISRGIYSCIHGVAIEERQSVMNSPTATMNNTHSNILRLLRTAKNMANLSGVNGSPQSALDFIRRESSVYDISEHRRSFTHSDCKSYNNPPCEENLFSDYISEVERTFGNLQLKDSNVYQDHYHHHHRPHSIGSASSIDGLYDCDNPPFTTQSRSISKKPLDIGLPSSKHSQLSDLYGKFSFKSDRYSGHDDLIRSDVSDISTHTVTYGNIEGNAAKRRKQQYKDSLKKRPASAKSRREFDEIELAYRRRPPRSPDHKRYFRDKEGLRDFYLDQFRTKENSPHWEHVDLTDIYKERSDDFKRDSVSGGGPCTNRSHIKHGTGDKHGVVSGVPAPWEKNLTNVEWEDRSGGNFCRSCPSKLHNYSTTVTGQNSGRQACIRCEACKKAGNLYDISEDNSLQELDQPAAPVAVTSNASTTKYPQSPTNSKAQKKNRNKLRRQHSYDTFVDLQKEEAALAPRSVSLKDKGRFMDGSPYAHMFEMSAGESTFANNKSSVPTAGHHHHNNPGGGYMLSKSLYPDRVTQNPFIPTFGDDQCLLHGSKSYFFRQPTVAGASKARPDFRALVTNKPVVSALHGAVPARFQKDICIGNQSNPCVPNNKNPRAFNGSSNGHVYEKLSSIESDV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q13224 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S31 | Phosphorylation | Uniprot | |
| S34 | Phosphorylation | Uniprot | |
| Y239 | Phosphorylation | Uniprot | |
| S383 | Phosphorylation | Q13554 (CAMK2B) , Q9UQM7 (CAMK2A) | Uniprot |
| T475 | Phosphorylation | Uniprot | |
| T493 | Phosphorylation | Uniprot | |
| T514 | Phosphorylation | Uniprot | |
| S720 | Phosphorylation | Uniprot | |
| S759 | Phosphorylation | Uniprot | |
| T760 | Phosphorylation | Uniprot | |
| Y762 | Phosphorylation | Uniprot | |
| S770 | Phosphorylation | Uniprot | |
| S865 | Phosphorylation | Uniprot | |
| Y932 | Phosphorylation | P06241 (FYN) | Uniprot |
| Y1039 | Phosphorylation | P06241 (FYN) | Uniprot |
| S1043 | Phosphorylation | Uniprot | |
| Y1070 | Phosphorylation | P06241 (FYN) | Uniprot |
| Y1109 | Phosphorylation | P06241 (FYN) | Uniprot |
| S1116 | Phosphorylation | Uniprot | |
| S1166 | Phosphorylation | Uniprot | |
| S1168 | Phosphorylation | Uniprot | |
| T1183 | Phosphorylation | Uniprot | |
| S1191 | Phosphorylation | Uniprot | |
| Y1252 | Phosphorylation | P06241 (FYN) | Uniprot |
| K1280 | Acetylation | Uniprot | |
| S1284 | Phosphorylation | Uniprot | |
| K1289 | Acetylation | Uniprot | |
| S1303 | Phosphorylation | P17252 (PRKCA) , P05771 (PRKCB) , Q13554 (CAMK2B) , P53355 (DAPK1) , P05129 (PRKCG) , Q9UQM7 (CAMK2A) | Uniprot |
| Y1304 | Phosphorylation | Uniprot | |
| T1306 | Phosphorylation | Uniprot | |
| S1323 | Phosphorylation | P17252 (PRKCA) , P05129 (PRKCG) , P05771 (PRKCB) | Uniprot |
| Y1336 | Phosphorylation | P06241 (FYN) | Uniprot |
| T1410 | Phosphorylation | Uniprot | |
| S1432 | Phosphorylation | Uniprot | |
| S1469 | Phosphorylation | Uniprot | |
| Y1474 | Phosphorylation | P29323 (EPHB2) , P12931 (SRC) , P06241 (FYN) | Uniprot |
| S1478 | Phosphorylation | Calculation | |
| S1479 | Phosphorylation | P68400 (CSNK2A1) | Uniprot |
| S1482 | Phosphorylation | Uniprot |
Research Backgrounds
Component of NMDA receptor complexes that function as heterotetrameric, ligand-gated ion channels with high calcium permeability and voltage-dependent sensitivity to magnesium. Channel activation requires binding of the neurotransmitter glutamate to the epsilon subunit, glycine binding to the zeta subunit, plus membrane depolarization to eliminate channel inhibition by Mg(2+). Sensitivity to glutamate and channel kinetics depend on the subunit composition. In concert with DAPK1 at extrasynaptic sites, acts as a central mediator for stroke damage. Its phosphorylation at Ser-1303 by DAPK1 enhances synaptic NMDA receptor channel activity inducing injurious Ca2+ influx through them, resulting in an irreversible neuronal death. Contributes to neural pattern formation in the developing brain. Plays a role in long-term depression (LTD) of hippocampus membrane currents and in synaptic plasticity (By similarity).
Phosphorylated on tyrosine residues (By similarity). Phosphorylation at Ser-1303 by DAPK1 enhances synaptic NMDA receptor channel activity (By similarity).
Cell membrane>Multi-pass membrane protein. Cell junction>Synapse>Postsynaptic cell membrane>Multi-pass membrane protein. Late endosome. Lysosome.
Primarily found in the fronto-parieto-temporal cortex and hippocampus pyramidal cells, lower expression in the basal ganglia.
Heterotetramer. Forms heterotetrameric channels composed of two zeta subunits (GRIN1), and two epsilon subunits (GRIN2A, GRIN2B, GRIN2C or GRIN2D) (in vitro). Can also form heterotetrameric channels that contain at least one zeta subunit (GRIN1), at least one epsilon subunit, plus GRIN3A or GRIN3B (By similarity). In vivo, the subunit composition may depend on the expression levels of the different subunits (Probable). Found in a complex with GRIN1 and GRIN3B. Found in a complex with GRIN1, GRIN3A and PPP2CB. Interacts with PDZ domains of PATJ, DLG3 and DLG4. Interacts with HIP1 and NETO1 (By similarity). Interacts with MAGI3. Interacts with DAPK1 (By similarity). Found in a complex with GRIN1 and PRR7. Interacts with PRR7. Interacts with CAMK2A. Interacts with ARC; preventing ARC oligomerization (By similarity). Interacts with TMEM25 (By similarity).
A hydrophobic region that gives rise to the prediction of a transmembrane span does not cross the membrane, but is part of a discontinuously helical region that dips into the membrane and is probably part of the pore and of the selectivity filter.
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family. NR2B/GRIN2B subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signaling molecules and interaction > Neuroactive ligand-receptor interaction.
· Human Diseases > Neurodegenerative diseases > Alzheimer's disease.
· Human Diseases > Neurodegenerative diseases > Amyotrophic lateral sclerosis (ALS).
· Human Diseases > Neurodegenerative diseases > Huntington's disease.
· Human Diseases > Substance dependence > Cocaine addiction.
· Human Diseases > Substance dependence > Amphetamine addiction.
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Immune diseases > Systemic lupus erythematosus.
· Organismal Systems > Environmental adaptation > Circadian entrainment.
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Glutamatergic synapse.
· Organismal Systems > Nervous system > Dopaminergic synapse.
References
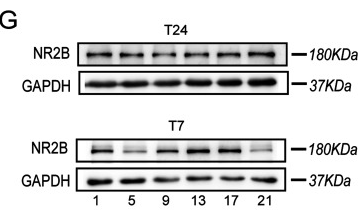
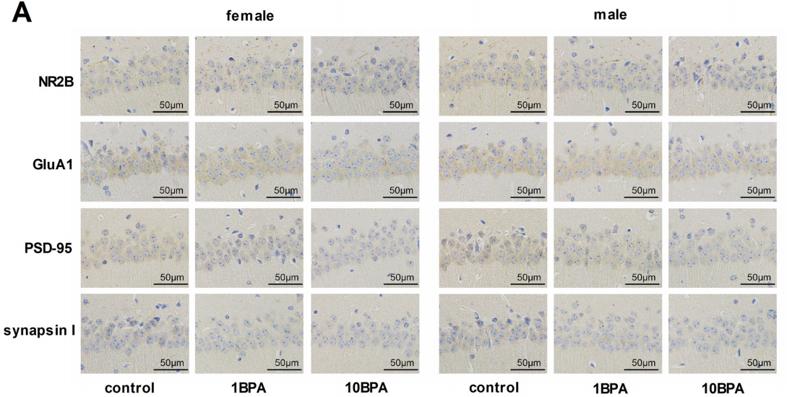
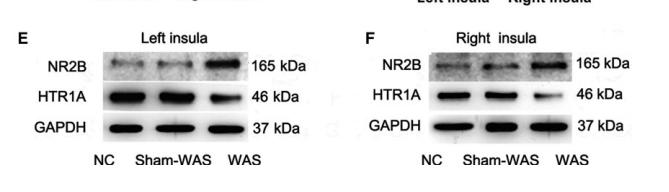
Application: WB Species: Mouse Sample:
Application: IHC Species: rat Sample: hippocampus
Application: WB Species: Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.