CDC25A Antibody - #AF6252
| Product: | CDC25A Antibody |
| Catalog: | AF6252 |
| Description: | Rabbit polyclonal antibody to CDC25A |
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 60kDa; 59kD(Calculated). |
| Uniprot: | P30304 |
| RRID: | AB_2835116 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6252, RRID:AB_2835116.
Fold/Unfold
Cdc 25a; CDC25A; CDC25A2; CDC25A2 CAG isoform; Cell division cycle 25 homolog A (S. pombe); Cell division cycle 25A; Cell division cycle 25A isoform a; Cell division cycle 25A isoform b; D9Ertd393e; Dual specificity phosphatase Cdc25A; M phase inducer phosphatase 1; M-phase inducer phosphatase 1; MGC115549; MPIP1_HUMAN;
Immunogens
- P30304 MPIP1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MELGPEPPHRRRLLFACSPPPASQPVVKALFGASAAGGLSPVTNLTVTMDQLQGLGSDYEQPLEVKNNSNLQRMGSSESTDSGFCLDSPGPLDSKENLENPMRRIHSLPQKLLGCSPALKRSHSDSLDHDIFQLIDPDENKENEAFEFKKPVRPVSRGCLHSHGLQEGKDLFTQRQNSAPARMLSSNERDSSEPGNFIPLFTPQSPVTATLSDEDDGFVDLLDGENLKNEEETPSCMASLWTAPLVMRTTNLDNRCKLFDSPSLCSSSTRSVLKRPERSQEESPPGSTKRRKSMSGASPKESTNPEKAHETLHQSLSLASSPKGTIENILDNDPRDLIGDFSKGYLFHTVAGKHQDLKYISPEIMASVLNGKFANLIKEFVIIDCRYPYEYEGGHIKGAVNLHMEEEVEDFLLKKPIVPTDGKRVIVVFHCEFSSERGPRMCRYVRERDRLGNEYPKLHYPELYVLKGGYKEFFMKCQSYCEPPSYRPMHHEDFKEDLKKFRTKSRTWAGEKSKREMYSRLKKL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P30304 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Ubiquitination | Uniprot | ||
| S18 | Phosphorylation | P06493 (CDK1) | Uniprot |
| S40 | Phosphorylation | Uniprot | |
| Y59 | Phosphorylation | Uniprot | |
| S76 | Phosphorylation | O14757 (CHEK1) , P49841 (GSK3B) | Uniprot |
| S77 | Phosphorylation | Uniprot | |
| S79 | Phosphorylation | Q8NG66 (NEK11) , P48729 (CSNK1A1) | Uniprot |
| T80 | Phosphorylation | Q9H4B4 (PLK3) | Uniprot |
| S82 | Phosphorylation | P48729 (CSNK1A1) , Q8NG66 (NEK11) | Uniprot |
| S88 | Phosphorylation | Q8NG66 (NEK11) | Uniprot |
| S107 | Phosphorylation | Uniprot | |
| K111 | Ubiquitination | Uniprot | |
| S116 | Phosphorylation | P06493 (CDK1) | Uniprot |
| K120 | Ubiquitination | Uniprot | |
| S124 | Phosphorylation | Q683Z8 (CHK2) , O96017 (CHEK2) , O14757 (CHEK1) | Uniprot |
| S126 | Phosphorylation | Uniprot | |
| K150 | Acetylation | Uniprot | |
| K150 | Ubiquitination | Uniprot | |
| K169 | Ubiquitination | Uniprot | |
| S178 | Phosphorylation | Q683Z8 (CHK2) , O14757 (CHEK1) , O96017 (CHEK2) | Uniprot |
| K257 | Ubiquitination | Uniprot | |
| S261 | Phosphorylation | Uniprot | |
| S263 | Phosphorylation | P24941 (CDK2) | Uniprot |
| R278 | Methylation | Uniprot | |
| S279 | Phosphorylation | Q683Z8 (CHK2) , O14757 (CHEK1) , O96017 (CHEK2) | Uniprot |
| S283 | Phosphorylation | Uniprot | |
| R290 | Methylation | Uniprot | |
| S293 | Phosphorylation | O14757 (CHEK1) , O96017 (CHEK2) , Q15418 (RPS6KA1) , Q683Z8 (CHK2) | Uniprot |
| S295 | Phosphorylation | Q15418 (RPS6KA1) | Uniprot |
| K300 | Ubiquitination | Uniprot | |
| S321 | Phosphorylation | Uniprot | |
| K343 | Ubiquitination | Uniprot | |
| K353 | Ubiquitination | Uniprot | |
| K358 | Ubiquitination | Uniprot | |
| Y359 | Phosphorylation | Uniprot | |
| S361 | Phosphorylation | Uniprot | |
| S367 | Phosphorylation | Uniprot | |
| K372 | Ubiquitination | Uniprot | |
| K378 | Ubiquitination | Uniprot | |
| K415 | Ubiquitination | Uniprot | |
| K471 | Ubiquitination | Uniprot | |
| Y486 | Phosphorylation | Uniprot | |
| T507 | Phosphorylation | O14757 (CHEK1) | Uniprot |
| S513 | Phosphorylation | Q9H4B4 (PLK3) | Uniprot |
| Y518 | Phosphorylation | Uniprot | |
| S519 | Phosphorylation | Q9H4B4 (PLK3) | Uniprot |
Research Backgrounds
Tyrosine protein phosphatase which functions as a dosage-dependent inducer of mitotic progression. Directly dephosphorylates CDK1 and stimulates its kinase activity. Also dephosphorylates CDK2 in complex with cyclin E, in vitro.
Phosphorylated by CHEK1 on Ser-76, Ser-124, Ser-178, Ser-279, Ser-293 and Thr-507 during checkpoint mediated cell cycle arrest. Also phosphorylated by CHEK2 on Ser-124, Ser-279, and Ser-293 during checkpoint mediated cell cycle arrest. Phosphorylation on Ser-178 and Thr-507 creates binding sites for YWHAE/14-3-3 epsilon which inhibits CDC25A. Phosphorylation on Ser-76, Ser-124, Ser-178, Ser-279 and Ser-293 may also promote ubiquitin-dependent proteolysis of CDC25A by the SCF complex. Phosphorylation of CDC25A at Ser-76 by CHEK1 primes it for subsequent phosphorylation at Ser-79, Ser-82 and Ser-88 by NEK11. Phosphorylation by NEK11 is required for BTRC-mediated polyubiquitination and degradation. Phosphorylation by PIM1 leads to an increase in phosphatase activity. Phosphorylated by PLK3 following DNA damage, leading to promote its ubiquitination and degradation.
Ubiquitinated by the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase complex that contains FZR1/CDH1 during G1 phase leading to its degradation by the proteasome. Ubiquitinated by a SCF complex containing BTRC and FBXW11 during S phase leading to its degradation by the proteasome. Deubiquitination by USP17L2/DUB3 leads to its stabilization.
Interacts with CCNB1/cyclin B1. Interacts with YWHAE/14-3-3 epsilon when phosphorylated. Interacts with CUL1 specifically when CUL1 is neddylated and active. Interacts with BTRC/BTRCP1 and FBXW11/BTRCP2. Interactions with CUL1, BTRC and FBXW11 are enhanced upon DNA damage. Interacts with PIM1. Interacts with CHEK2; mediates CDC25A phosphorylation and degradation in response to infrared-induced DNA damages. Interacts with HSP90AB1; prevents heat shock-mediated CDC25A degradation and contributes to cell cycle progression.
The phosphodegron motif mediates interaction with specific F-box proteins when phosphorylated. Putative phosphorylation sites at Ser-79 and Ser-82 appear to be essential for this interaction.
Belongs to the MPI phosphatase family.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
References
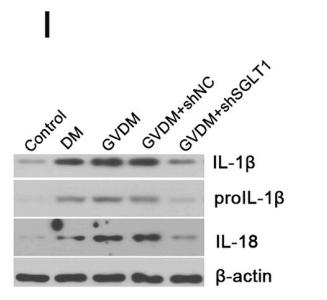
Application: WB Species: mice Sample: left ventricle tissues
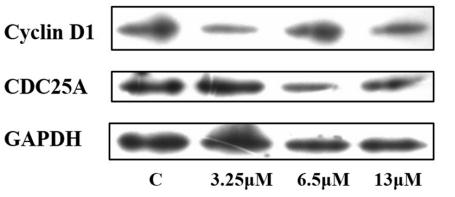
Application: WB Species: human Sample: A549 cells
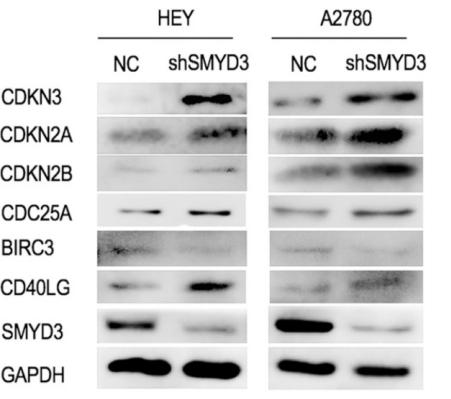
Application: WB Species: human Sample: HEY and A2780 cells
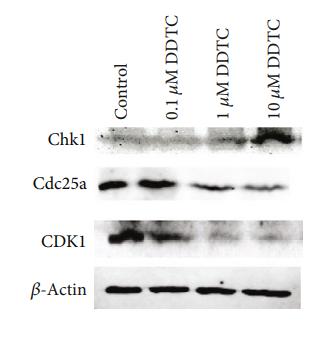
Application: WB Species: human Sample: SKOV3 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.