Histone H2A.X Antibody - #AF6187
| Product: | Histone H2A.X Antibody |
| Catalog: | AF6187 |
| Description: | Rabbit polyclonal antibody to Histone H2A.X |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Bovine, Sheep, Dog |
| Mol.Wt.: | 15kDa; 15kD(Calculated). |
| Uniprot: | P16104 |
| RRID: | AB_2835070 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6187, RRID:AB_2835070.
Fold/Unfold
AW228881; H2A histone family member X; H2A.FX; H2A.X; H2a/x; H2AFX; H2AX; H2AX histone; H2AX_HUMAN; Hist5.2ax; Histone 2A; Histone 2AX; Histone H2A.X; Histone H2AX; RGD1566119;
Immunogens
- P16104 H2AX_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSGRGKTGGKARAKAKSRSSRAGLQFPVGRVHRLLRKGHYAERVGAGAPVYLAAVLEYLTAEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGGVTIAQGGVLPNIQAVLLPKKTSATVGPKAPSGGKKATQASQEY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P16104 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S2 | Phosphorylation | Uniprot | |
| K6 | Acetylation | Uniprot | |
| K10 | Acetylation | Uniprot | |
| K14 | Ubiquitination | Uniprot | |
| K16 | Ubiquitination | Uniprot | |
| S19 | Phosphorylation | Uniprot | |
| K119 | Acetylation | Uniprot | |
| K119 | Ubiquitination | Uniprot | |
| K120 | Ubiquitination | Uniprot | |
| T121 | Phosphorylation | Uniprot | |
| S122 | Phosphorylation | Uniprot | |
| T124 | Phosphorylation | Uniprot | |
| K134 | Methylation | Uniprot | |
| K134 | Sumoylation | Uniprot | |
| K134 | Ubiquitination | Uniprot | |
| T137 | Phosphorylation | Uniprot | |
| S140 | Phosphorylation | Q13315 (ATM) , P45984 (MAPK9) , P45983 (MAPK8) | Uniprot |
| Y143 | Phosphorylation | Q9UIG0 (BAZ1B) | Uniprot |
Research Backgrounds
Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation.
Phosphorylated on Ser-140 (to form gamma-H2AX or H2AX139ph) in response to DNA double strand breaks (DSBs) generated by exogenous genotoxic agents and by stalled replication forks, and may also occur during meiotic recombination events and immunoglobulin class switching in lymphocytes. Phosphorylation can extend up to several thousand nucleosomes from the actual site of the DSB and may mark the surrounding chromatin for recruitment of proteins required for DNA damage signaling and repair. Widespread phosphorylation may also serve to amplify the damage signal or aid repair of persistent lesions. Phosphorylation of Ser-140 (H2AX139ph) in response to ionizing radiation is mediated by both ATM and PRKDC while defects in DNA replication induce Ser-140 phosphorylation (H2AX139ph) subsequent to activation of ATR and PRKDC. Dephosphorylation of Ser-140 by PP2A is required for DNA DSB repair. In meiosis, Ser-140 phosphorylation (H2AX139ph) may occur at synaptonemal complexes during leptotene as an ATM-dependent response to the formation of programmed DSBs by SPO11. Ser-140 phosphorylation (H2AX139ph) may subsequently occurs at unsynapsed regions of both autosomes and the XY bivalent during zygotene, downstream of ATR and BRCA1 activation. Ser-140 phosphorylation (H2AX139ph) may also be required for transcriptional repression of unsynapsed chromatin and meiotic sex chromosome inactivation (MSCI), whereby the X and Y chromosomes condense in pachytene to form the heterochromatic XY-body. During immunoglobulin class switch recombination in lymphocytes, Ser-140 phosphorylation (H2AX139ph) may occur at sites of DNA-recombination subsequent to activation of the activation-induced cytidine deaminase AICDA. Phosphorylation at Tyr-143 (H2AXY142ph) by BAZ1B/WSTF determines the relative recruitment of either DNA repair or pro-apoptotic factors. Phosphorylation at Tyr-143 (H2AXY142ph) favors the recruitment of APBB1/FE65 and pro-apoptosis factors such as MAPK8/JNK1, triggering apoptosis. In contrast, dephosphorylation of Tyr-143 by EYA proteins (EYA1, EYA2, EYA3 or EYA4) favors the recruitment of MDC1-containing DNA repair complexes to the tail of phosphorylated Ser-140 (H2AX139ph).
Monoubiquitination of Lys-120 (H2AXK119ub) by RING1 and RNF2/RING2 complex gives a specific tag for epigenetic transcriptional repression (By similarity). Following DNA double-strand breaks (DSBs), it is ubiquitinated through 'Lys-63' linkage of ubiquitin moieties by the E2 ligase UBE2N and the E3 ligases RNF8 and RNF168, leading to the recruitment of repair proteins to sites of DNA damage. Ubiquitination at Lys-14 and Lys-16 (H2AK13Ub and H2AK15Ub, respectively) in response to DNA damage is initiated by RNF168 that mediates monoubiquitination at these 2 sites, and 'Lys-63'-linked ubiquitin are then conjugated to monoubiquitin; RNF8 is able to extend 'Lys-63'-linked ubiquitin chains in vitro. H2AK119Ub and ionizing radiation-induced 'Lys-63'-linked ubiquitination (H2AK13Ub and H2AK15Ub) are distinct events.
Acetylation at Lys-37 increases in S and G2 phases. This modification has been proposed to play a role in DNA double-strand break repair (By similarity).
Nucleus. Chromosome.
The nucleosome is a histone octamer containing two molecules each of H2A, H2B, H3 and H4 assembled in one H3-H4 heterotetramer and two H2A-H2B heterodimers. The octamer wraps approximately 147 bp of DNA (Probable). Interacts with numerous proteins required for DNA damage signaling and repair when phosphorylated on Ser-140. These include MDC1, TP53BP1, BRCA1 and the MRN complex, composed of MRE11, RAD50, and NBN. Interaction with the MRN complex is mediated at least in part by NBN. Also interacts with DHX9/NDHII when phosphorylated on Ser-140 and MCPH1 when phosphorylated at Ser-140 or Tyr-143. Interacts with ARRB2; the interaction is detected in the nucleus upon OR1D2 stimulation. Interacts with WRAP53/TCAB1.
(Microbial infection) Interacts with Epstein-Barr virus protein EBNA6.
The [ST]-Q motif constitutes a recognition sequence for kinases from the PI3/PI4-kinase family.
Belongs to the histone H2A family.
Research Fields
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Immune diseases > Systemic lupus erythematosus.
References
Application: WB Species: mouse Sample: lung
Application: WB Species: Mice Sample: THLE-2 cells
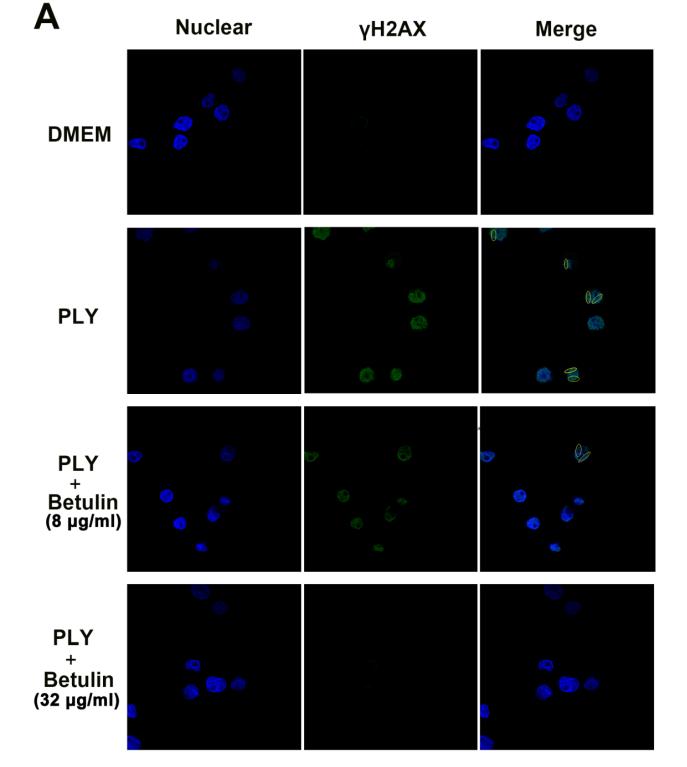
Application: IF/ICC Species: Human Sample: A549 cells
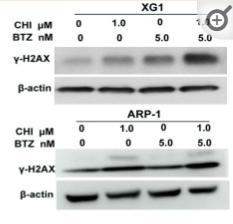
Application: WB Species: Human Sample: MM cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.