ATP1A1 Antibody - #AF6109
| Product: | ATP1A1 Antibody |
| Catalog: | AF6109 |
| Description: | Rabbit polyclonal antibody to ATP1A1 |
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 113kDa; 113kD(Calculated). |
| Uniprot: | P05023 |
| RRID: | AB_2834996 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6109, RRID:AB_2834996.
Fold/Unfold
A1A1; AT1A1; AT1A1_HUMAN; ATP1A1; Atpa-1; ATPase Na+/K+ transporting alpha 1 polypeptide; ATPase Na+/K+ transporting subunit alpha 1; BC010319; EC 3.6.3.9; MGC3285; MGC38419; MGC51750; Na K ATPase alpha A catalytic polypeptide; Na K ATPase catalytic subunit alpha A protein; Na(+)/K(+) ATPase 1; Na(+)/K(+) ATPase alpha-1 subunit; Na+; K+ ATPase alpha subunit; Na+/K+ ATPase alpha 1 subunit; Na+/K+ ATPase 1; Na;K ATPase alpha 1 subunit; Nkaa1b; Sodium potassium ATPase alpha 1 polypeptide; Sodium pump 1; Sodium pump subunit alpha-1; sodium-potassium ATPase catalytic subunit alpha-1; Sodium/potassium-transporting ATPase subunit alpha-1;
Immunogens
- P05023 AT1A1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGKGVGRDKYEPAAVSEQGDKKGKKGKKDRDMDELKKEVSMDDHKLSLDELHRKYGTDLSRGLTSARAAEILARDGPNALTPPPTTPEWIKFCRQLFGGFSMLLWIGAILCFLAYSIQAATEEEPQNDNLYLGVVLSAVVIITGCFSYYQEAKSSKIMESFKNMVPQQALVIRNGEKMSINAEEVVVGDLVEVKGGDRIPADLRIISANGCKVDNSSLTGESEPQTRSPDFTNENPLETRNIAFFSTNCVEGTARGIVVYTGDRTVMGRIATLASGLEGGQTPIAAEIEHFIHIITGVAVFLGVSFFILSLILEYTWLEAVIFLIGIIVANVPEGLLATVTVCLTLTAKRMARKNCLVKNLEAVETLGSTSTICSDKTGTLTQNRMTVAHMWFDNQIHEADTTENQSGVSFDKTSATWLALSRIAGLCNRAVFQANQENLPILKRAVAGDASESALLKCIELCCGSVKEMRERYAKIVEIPFNSTNKYQLSIHKNPNTSEPQHLLVMKGAPERILDRCSSILLHGKEQPLDEELKDAFQNAYLELGGLGERVLGFCHLFLPDEQFPEGFQFDTDDVNFPIDNLCFVGLISMIDPPRAAVPDAVGKCRSAGIKVIMVTGDHPITAKAIAKGVGIISEGNETVEDIAARLNIPVSQVNPRDAKACVVHGSDLKDMTSEQLDDILKYHTEIVFARTSPQQKLIIVEGCQRQGAIVAVTGDGVNDSPALKKADIGVAMGIAGSDVSKQAADMILLDDNFASIVTGVEEGRLIFDNLKKSIAYTLTSNIPEITPFLIFIIANIPLPLGTVTILCIDLGTDMVPAISLAYEQAESDIMKRQPRNPKTDKLVNERLISMAYGQIGMIQALGGFFTYFVILAENGFLPIHLLGLRVDWDDRWINDVEDSYGQQWTYEQRKIVEFTCHTAFFVSIVVVQWADLVICKTRRNSVFQQGMKNKILIFGLFEETALAAFLSYCPGMGVALRMYPLKPTWWFCAFPYSLLIFVYDEVRKLIIRRRPGGWVEKETYY
PTMs - P05023 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K9 | Ubiquitination | Uniprot | |
| Y10 | Phosphorylation | Uniprot | |
| S16 | Phosphorylation | Q05513 (PRKCZ) , P28482 (MAPK1) | Uniprot |
| K21 | Ubiquitination | Uniprot | |
| K36 | Ubiquitination | Uniprot | |
| K37 | Ubiquitination | Uniprot | |
| K45 | Ubiquitination | Uniprot | |
| S47 | Phosphorylation | Uniprot | |
| K54 | Ubiquitination | Uniprot | |
| Y55 | Phosphorylation | Uniprot | |
| S60 | Phosphorylation | Uniprot | |
| R61 | Methylation | Uniprot | |
| T81 | Phosphorylation | Uniprot | |
| T85 | Phosphorylation | Uniprot | |
| T86 | Phosphorylation | Uniprot | |
| Y149 | Phosphorylation | Uniprot | |
| K156 | Ubiquitination | Uniprot | |
| K162 | Ubiquitination | Uniprot | |
| K177 | Ubiquitination | Uniprot | |
| K194 | Ubiquitination | Uniprot | |
| R198 | Methylation | Uniprot | |
| R204 | Methylation | Uniprot | |
| S207 | Phosphorylation | Uniprot | |
| K212 | Ubiquitination | Uniprot | |
| S216 | Phosphorylation | Uniprot | |
| S217 | Phosphorylation | Uniprot | |
| S222 | Phosphorylation | Uniprot | |
| T226 | Phosphorylation | Uniprot | |
| S228 | Phosphorylation | Uniprot | |
| S246 | Phosphorylation | Uniprot | |
| C249 | S-Nitrosylation | Uniprot | |
| Y260 | Phosphorylation | Uniprot | |
| T261 | Phosphorylation | Uniprot | |
| R264 | Methylation | Uniprot | |
| T265 | Phosphorylation | Uniprot | |
| K359 | Ubiquitination | Uniprot | |
| S369 | Phosphorylation | Uniprot | |
| K377 | Ubiquitination | Uniprot | |
| T378 | Phosphorylation | Uniprot | |
| S407 | Phosphorylation | Uniprot | |
| K444 | Ubiquitination | Uniprot | |
| S452 | Phosphorylation | Uniprot | |
| K458 | Ubiquitination | Uniprot | |
| C459 | S-Nitrosylation | Uniprot | |
| C463 | S-Nitrosylation | Uniprot | |
| S466 | Phosphorylation | Uniprot | |
| K468 | Ubiquitination | Uniprot | |
| K476 | Ubiquitination | Uniprot | |
| S484 | Phosphorylation | Uniprot | |
| K487 | Acetylation | Uniprot | |
| K487 | Ubiquitination | Uniprot | |
| Y488 | Phosphorylation | Uniprot | |
| K494 | Methylation | Uniprot | |
| K494 | Ubiquitination | Uniprot | |
| T498 | Phosphorylation | Uniprot | |
| S499 | Phosphorylation | Uniprot | |
| K508 | Ubiquitination | Uniprot | |
| R517 | Methylation | Uniprot | |
| S520 | Phosphorylation | Uniprot | |
| K526 | Sumoylation | Uniprot | |
| K526 | Ubiquitination | Uniprot | |
| K535 | Ubiquitination | Uniprot | |
| Y542 | Phosphorylation | Uniprot | |
| K605 | Ubiquitination | Uniprot | |
| K612 | Ubiquitination | Uniprot | |
| T617 | Phosphorylation | Uniprot | |
| T623 | Phosphorylation | Uniprot | |
| K625 | Ubiquitination | Uniprot | |
| K629 | Ubiquitination | Uniprot | |
| T640 | Phosphorylation | Uniprot | |
| S653 | Phosphorylation | Uniprot | |
| K661 | Ubiquitination | Uniprot | |
| K671 | Ubiquitination | Uniprot | |
| K683 | Ubiquitination | Uniprot | |
| Y684 | Phosphorylation | Uniprot | |
| T686 | Phosphorylation | Uniprot | |
| K698 | Ubiquitination | Uniprot | |
| C705 | S-Nitrosylation | Uniprot | |
| S722 | Phosphorylation | Uniprot | |
| K726 | Ubiquitination | Uniprot | |
| K727 | Ubiquitination | Uniprot | |
| K773 | Ubiquitination | Uniprot | |
| K833 | Ubiquitination | Uniprot | |
| K840 | Ubiquitination | Uniprot | |
| K843 | Ubiquitination | Uniprot | |
| Y902 | Phosphorylation | Uniprot | |
| S943 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K950 | Ubiquitination | Uniprot | |
| K952 | Ubiquitination | Uniprot | |
| K1019 | Ubiquitination | Uniprot |
Research Backgrounds
This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
Phosphorylation on Tyr-10 modulates pumping activity. Phosphorylation of Ser-943 by PKA modulates the response of ATP1A1 to PKC. Dephosphorylation by protein phosphatase 2A (PP2A) following increases in intracellular sodium, leading to increase catalytic activity (By similarity).
Cell membrane>Sarcolemma>Multi-pass membrane protein. Melanosome.
Note: Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
The sodium/potassium-transporting ATPase is composed of a catalytic alpha subunit, an auxiliary non-catalytic beta subunit and an additional regulatory subunit. Interacts with regulatory subunit FXYD1 (By similarity). Interacts with regulatory subunit FXYD3. Interacts with SIK1 (By similarity). Binds the HLA class II histocompatibility antigen DR1. Interacts with SLC35G1 and STIM1.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Organismal Systems > Circulatory system > Cardiac muscle contraction. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Endocrine system > Insulin secretion. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone synthesis.
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Aldosterone synthesis and secretion.
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
· Organismal Systems > Excretory system > Endocrine and other factor-regulated calcium reabsorption.
· Organismal Systems > Excretory system > Proximal tubule bicarbonate reclamation.
· Organismal Systems > Digestive system > Salivary secretion.
· Organismal Systems > Digestive system > Gastric acid secretion.
· Organismal Systems > Digestive system > Pancreatic secretion.
· Organismal Systems > Digestive system > Carbohydrate digestion and absorption.
· Organismal Systems > Digestive system > Protein digestion and absorption.
· Organismal Systems > Digestive system > Mineral absorption.
References
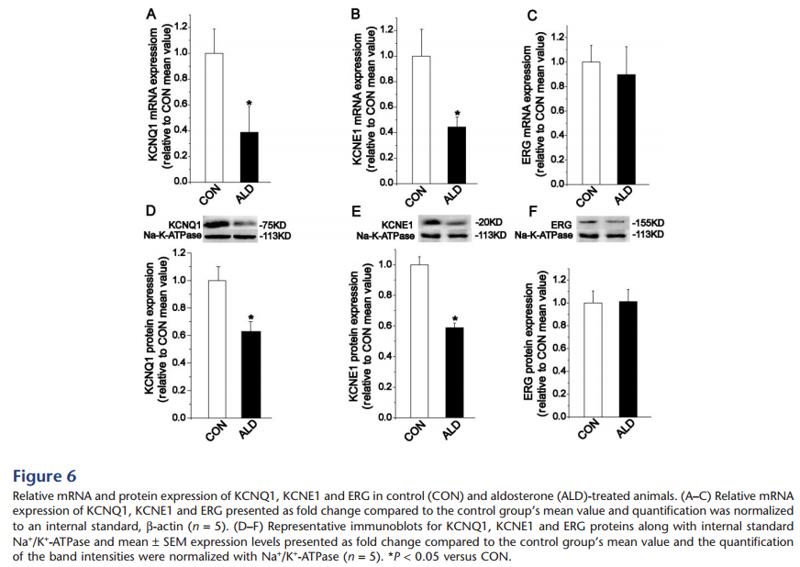
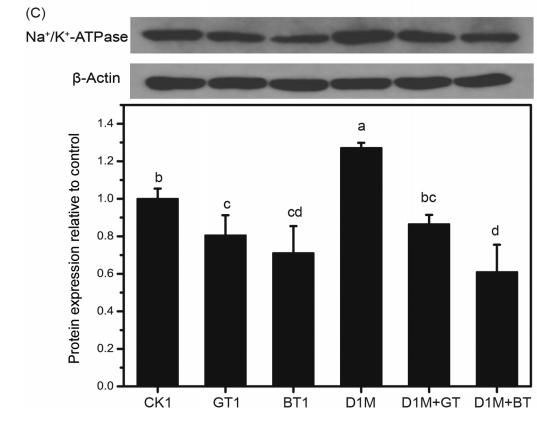
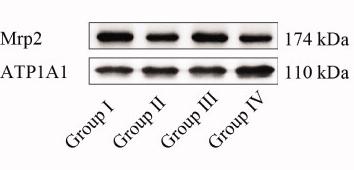
Application: WB Species: Mice Sample: heart tissues
Application: WB Species: mouse Sample: intestine
Application: WB Species: Rat Sample: ileum
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.