Estrogen Receptor-alpha Antibody - #AF6058
| Product: | Estrogen Receptor-alpha Antibody |
| Catalog: | AF6058 |
| Description: | Rabbit polyclonal antibody to Estrogen Receptor-alpha |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 66kDa,55kDa; 66kD(Calculated). |
| Uniprot: | P03372 |
| RRID: | AB_2834976 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6058, RRID:AB_2834976.
Fold/Unfold
DKFZp686N23123; ER alpha; ER; ER-alpha; Era; ESR; ESR1; ESR1_HUMAN; ESRA; Estradiol receptor; Estrogen nuclear receptor alpha; Estrogen receptor 1; Estrogen receptor alpha 3*,4,5,6,7*/822 isoform; Estrogen receptor alpha; Estrogen receptor alpha delta 3*,4,5,6,7*,8*/941 isoform; Estrogen receptor alpha delta 3*,4,5,6,7*/819 2 isoform; Estrogen receptor alpha delta 4 +49 isoform; Estrogen receptor alpha delta 4*,5,6,7*/654 isoform; Estrogen receptor alpha delta 4*,5,6,7,8*/901 isoform; Estrogen receptor alpha E1 E2 1 2; Estrogen receptor alpha E1 N2 E2 1 2; Estrogen receptor; ESTRR; NR3A1; Nuclear receptor subfamily 3 group A member 1;
Immunogens
- P03372 ESR1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTMTLHTKASGMALLHQIQGNELEPLNRPQLKIPLERPLGEVYLDSSKPAVYNYPEGAAYEFNAAAAANAQVYGQTGLPYGPGSEAAAFGSNGLGGFPPLNSVSPSPLMLLHPPPQLSPFLQPHGQQVPYYLENEPSGYTVREAGPPAFYRPNSDNRRQGGRERLASTNDKGSMAMESAKETRYCAVCNDYASGYHYGVWSCEGCKAFFKRSIQGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAANLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLLECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGASVEETDQSHLATAGSTSSHSLQKYYITGEAEGFPATV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P03372 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| T4 | Phosphorylation | Uniprot | |
| T7 | Phosphorylation | Uniprot | |
| S10 | Phosphorylation | Uniprot | |
| K32 | Acetylation | Uniprot | |
| S46 | Phosphorylation | Uniprot | |
| S47 | Phosphorylation | Uniprot | |
| Y52 | Phosphorylation | P00519 (ABL1) | Uniprot |
| S102 | Phosphorylation | P49841 (GSK3B) | Uniprot |
| S104 | Phosphorylation | P49841 (GSK3B) , P27361 (MAPK3) , P28482 (MAPK1) , P31749 (AKT1) , P24941 (CDK2) , Q00526 (CDK3) , P42345 (MTOR) | Uniprot |
| S106 | Phosphorylation | P24941 (CDK2) , Q00526 (CDK3) , P49841 (GSK3B) , P28482 (MAPK1) , P27361 (MAPK3) , P42345 (MTOR) , P31749 (AKT1) | Uniprot |
| S118 | Phosphorylation | P24941 (CDK2) , P27361 (MAPK3) , O15111 (CHUK) , P49841 (GSK3B) , P31749 (AKT1) , Q16539 (MAPK14) , P50613 (CDK7) , Q00526 (CDK3) , P28482 (MAPK1) | Uniprot |
| S154 | Phosphorylation | Uniprot | |
| S167 | Phosphorylation | O14965 (AURKA) , P68400 (CSNK2A1) , P51812 (RPS6KA3) , P31751 (AKT2) , P23443 (RPS6KB1) , Q14164 (IKBKE) , P31749 (AKT1) , Q15418 (RPS6KA1) | Uniprot |
| K171 | Acetylation | Uniprot | |
| K171 | Methylation | Uniprot | |
| K171 | Sumoylation | Uniprot | |
| K180 | Methylation | Uniprot | |
| K180 | Sumoylation | Uniprot | |
| S212 | Phosphorylation | Uniprot | |
| Y219 | Phosphorylation | P00519 (ABL1) | Uniprot |
| S236 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K244 | Acetylation | Uniprot | |
| K252 | Acetylation | Uniprot | |
| R260 | Methylation | Uniprot | |
| K266 | Acetylation | Uniprot | |
| K266 | Methylation | Uniprot | |
| K266 | Sumoylation | Uniprot | |
| K268 | Acetylation | Uniprot | |
| K268 | Sumoylation | Uniprot | |
| R277 | Methylation | Uniprot | |
| S282 | Phosphorylation | P68400 (CSNK2A1) | Uniprot |
| S294 | Phosphorylation | P24941 (CDK2) , Q16539 (MAPK14) | Uniprot |
| K299 | Acetylation | Uniprot | |
| K299 | Sumoylation | Uniprot | |
| K302 | Acetylation | Uniprot | |
| K302 | Methylation | Uniprot | |
| K302 | Ubiquitination | Uniprot | |
| K303 | Acetylation | Uniprot | |
| S305 | Phosphorylation | Q13153 (PAK1) , P31749 (AKT1) , P17612 (PRKACA) , O14965 (AURKA) , Q9UHD2 (TBK1) , O96013 (PAK4) | Uniprot |
| T311 | Phosphorylation | Q16539 (MAPK14) | Uniprot |
| S341 | Phosphorylation | Uniprot | |
| K401 | Methylation | Uniprot | |
| K472 | Acetylation | Uniprot | |
| K472 | Sumoylation | Uniprot | |
| T485 | Phosphorylation | Uniprot | |
| Y537 | Phosphorylation | P06239 (LCK) , P12931 (SRC) | Uniprot |
| S554 | Phosphorylation | Uniprot | |
| S559 | Phosphorylation | P68400 (CSNK2A1) | Uniprot |
| S576 | Phosphorylation | Uniprot | |
| S578 | Phosphorylation | Uniprot | |
| T594 | Phosphorylation | Uniprot |
Research Backgrounds
Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full-length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1.
Phosphorylated by cyclin A/CDK2 and CK1. Phosphorylation probably enhances transcriptional activity. Self-association induces phosphorylation. Dephosphorylation at Ser-118 by PPP5C inhibits its transactivation activity. Phosphorylated by LMTK3 in vitro.
Glycosylated; contains N-acetylglucosamine, probably O-linked.
Ubiquitinated; regulated by LATS1 via DCAF1 it leads to ESR1 proteasomal degradation. Deubiquitinated by OTUB1.
Dimethylated by PRMT1 at Arg-260. The methylation may favor cytoplasmic localization. Demethylated by JMJD6 at Arg-260.
Palmitoylated (isoform 3). Not biotinylated (isoform 3).
Palmitoylated by ZDHHC7 and ZDHHC21. Palmitoylation is required for plasma membrane targeting and for rapid intracellular signaling via ERK and AKT kinases and cAMP generation, but not for signaling mediated by the nuclear hormone receptor.
Nucleus. Cytoplasm. Cell membrane>Peripheral membrane protein>Cytoplasmic side.
Note: A minor fraction is associated with the inner membrane.
Nucleus. Cytoplasm. Cell membrane>Peripheral membrane protein>Cytoplasmic side. Cell membrane>Single-pass type I membrane protein.
Note: Associated with the inner membrane via palmitoylation (Probable). At least a subset exists as a transmembrane protein with a N-terminal extracellular domain.
Nucleus. Golgi apparatus. Cell membrane.
Note: Colocalizes with ZDHHC7 and ZDHHC21 in the Golgi apparatus where most probably palmitoylation occurs. Associated with the plasma membrane when palmitoylated.
Widely expressed. Isoform 3 is not expressed in the pituitary gland.
Binds DNA as a homodimer. Can form a heterodimer with ESR2. Isoform 3 can probably homodimerize or heterodimerize with isoform 1 and ESR2. Interacts with FOXC2, MAP1S, SLC30A9, UBE1C and NCOA3 coactivator (By similarity). Interacts with EP300; the interaction is estrogen-dependent and enhanced by CITED1. Interacts with CITED1; the interaction is estrogen-dependent. Interacts with NCOA5 and NCOA6 coactivators. Interacts with NCOA7; the interaction is a ligand-inducible. Interacts with PHB2, PELP1 and UBE1C. Interacts with AKAP13. Interacts with CUEDC2. Interacts with KDM5A. Interacts with SMARD1. Interacts with HEXIM1. Interacts with PBXIP1. Interaction with MUC1 is stimulated by 7 beta-estradiol (E2) and enhances ERS1-mediated transcription. Interacts with DNTTIP2, FAM120B and UIMC1. Interacts with isoform 4 of TXNRD1. Interacts with KMT2D/MLL2. Interacts with ATAD2 and this interaction is enhanced by estradiol. Interacts with KIF18A and LDB1. Interacts with RLIM (via C-terminus). Interacts with MACROD1. Interacts with SH2D4A and PLCG. Interaction with SH2D4A blocks binding to PLCG and inhibits estrogen-induced cell proliferation. Interacts with DYNLL1. Interacts with CCDC62 in the presence of estradiol/E2; this interaction seems to enhance the transcription of target genes. Interacts with NR2C1; the interaction prevents homodimerization of ESR1 and suppresses its transcriptional activity and cell growth. Interacts with DNAAF4. Interacts with PRMT2. Interacts with PI3KR1 or PI3KR2, SRC and PTK2/FAK1. Interacts with RBFOX2. Interacts with STK3/MST2 only in the presence of SAV1 and vice-versa. Binds to CSNK1D. Interacts with NCOA2; NCOA2 can interact with ESR1 AF-1 and AF-2 domains simultaneously and mediate their transcriptional synergy. Interacts with DDX5. Interacts with NCOA1; the interaction seems to require a self-association of N-terminal and C-terminal regions. Interacts with ZNF366, DDX17, NFKB1, RELA, SP1 and SP3. Interacts with NRIP1 (By similarity). Interacts with GPER1; the interaction occurs in an estrogen-dependent manner. Interacts with CLOCK and the interaction is stimulated by estrogen. Interacts with BCAS3. Interacts with TRIP4 (ufmylated); estrogen dependent. Interacts with LMTK3; the interaction phosphorylates ESR1 (in vitro) and protects it against proteasomal degradation. Interacts with CCAR2 (via N-terminus) in a ligand-independent manner. Interacts with ZFHX3. Interacts with SFR1 in a ligand-dependent and -independent manner. Interacts with DCAF13, LATS1 and DCAF1; regulates ESR1 ubiquitination and ubiquitin-mediated proteasomal degradation. Interacts (via DNA-binding domain) with POU4F2 (C-terminus); this interaction increases the estrogen receptor ESR1 transcriptional activity in a DNA- and ligand 17-beta-estradiol-independent manner (By similarity). Interacts with ESRRB isoform 1. Interacts with UBE3A and WBP2. Interacts with GTF2B. Interacts with RBM39 (By similarity). In the absence of hormonal ligand, interacts with TACC1.
Composed of three domains: a modulating N-terminal domain, a DNA-binding domain and a C-terminal ligand-binding domain. The modulating domain, also known as A/B or AF-1 domain has a ligand-independent transactivation function. The C-terminus contains a ligand-dependent transactivation domain, also known as E/F or AF-2 domain which overlaps with the ligand binding domain. AF-1 and AF-2 activate transcription independently and synergistically and act in a promoter- and cell-specific manner. AF-1 seems to provide the major transactivation function in differentiated cells.
Belongs to the nuclear hormone receptor family. NR3 subfamily.
Research Fields
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Excretory system > Endocrine and other factor-regulated calcium reabsorption.
References
Application: IHC Species: rat Sample: thyroid
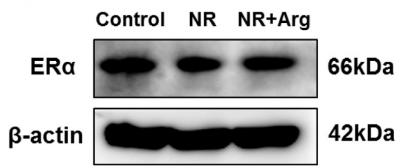
Application: WB Species: Human Sample: HepG2 cells
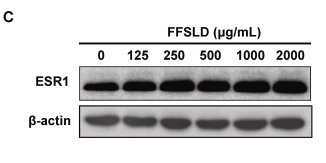
Application: WB Species: Human Sample:
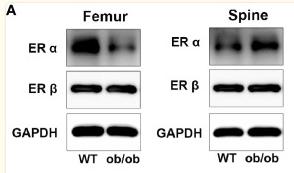
Application: WB Species: Mice Sample: femoral and spine
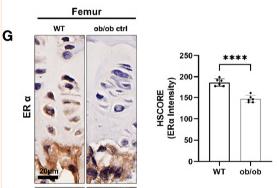
Application: IHC Species: Mice Sample: femoral and vertebral
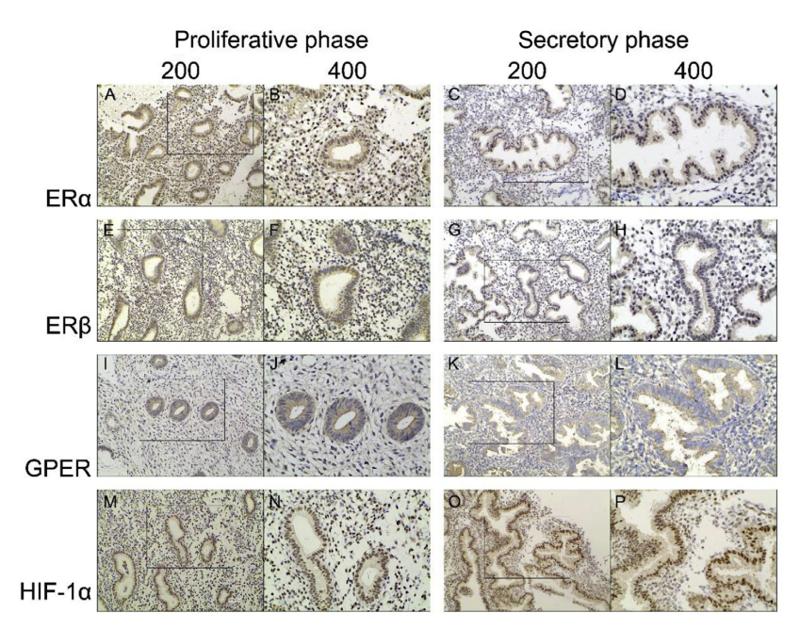
Application: IHC Species: human Sample: endometrium
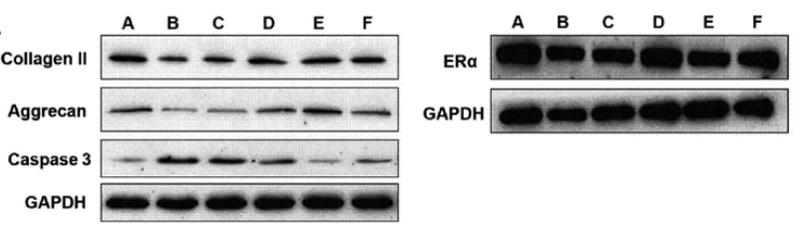
Application: WB Species: human Sample: chondrocytes
Application: WB Species: sheep Sample: endometrium
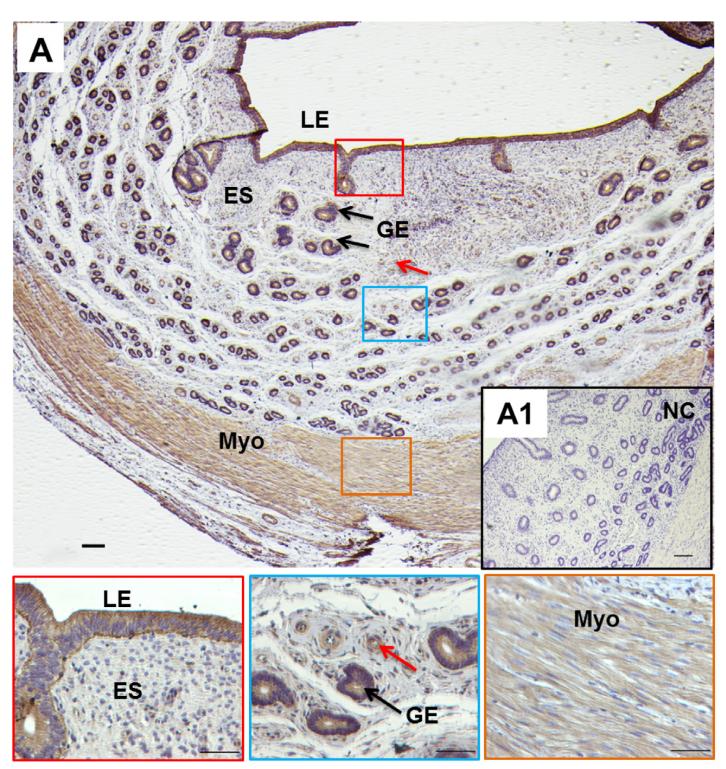
Application: IHC Species: sheep Sample: uterus
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.