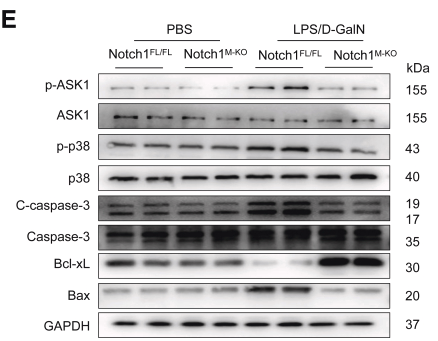
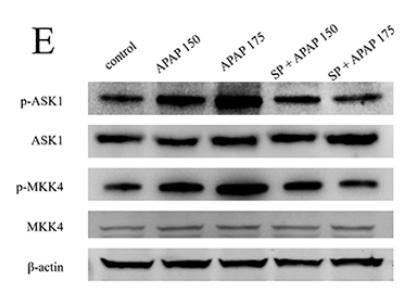
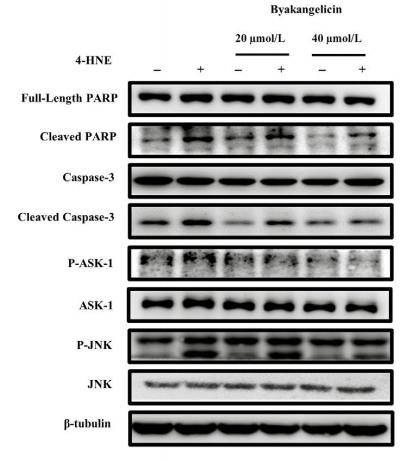
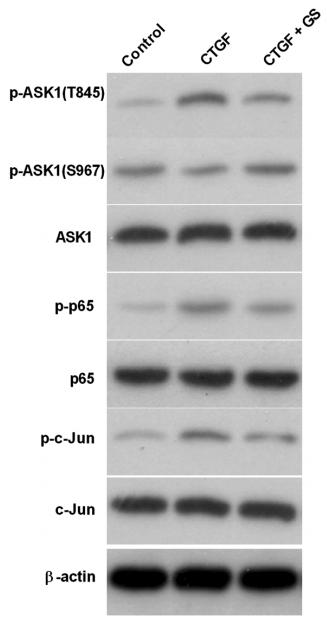
Phospho-ASK1 (Ser966) Antibody - #AF3477
| Product: | Phospho-ASK1 (Ser966) Antibody |
| Catalog: | AF3477 |
| Description: | Rabbit polyclonal antibody to Phospho-ASK1 (Ser966) |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 155kDa; 155kD(Calculated). |
| Uniprot: | Q99683 |
| RRID: | AB_2834915 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3477, RRID:AB_2834915.
Fold/Unfold
Apoptosis signal regulating kinase 1; Apoptosis signal-regulating kinase 1; ASK 1; ASK-1; ASK1; M3K5; M3K5_HUMAN; MAP/ERK kinase kinase 5; MAP3K5; MAPK/ERK kinase kinase 5; MAPKKK5; MEK kinase 5; MEKK 5; MEKK5; Mitogen activated protein kinase kinase kinase 5; Mitogen-activated protein kinase kinase kinase 5;
Immunogens
- Q99683 M3K5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSTEADEGITFSVPPFAPSGFCTIPEGGICRRGGAAAVGEGEEHQLPPPPPGSFWNVESAAAPGIGCPAATSSSSATRGRGSSVGGGSRRTTVAYVINEASQGQLVVAESEALQSLREACETVGATLETLHFGKLDFGETTVLDRFYNADIAVVEMSDAFRQPSLFYHLGVRESFSMANNIILYCDTNSDSLQSLKEIICQKNTMCTGNYTFVPYMITPHNKVYCCDSSFMKGLTELMQPNFELLLGPICLPLVDRFIQLLKVAQASSSQYFRESILNDIRKARNLYTGKELAAELARIRQRVDNIEVLTADIVINLLLSYRDIQDYDSIVKLVETLEKLPTFDLASHHHVKFHYAFALNRRNLPGDRAKALDIMIPMVQSEGQVASDMYCLVGRIYKDMFLDSNFTDTESRDHGASWFKKAFESEPTLQSGINYAVLLLAAGHQFESSFELRKVGVKLSSLLGKKGNLEKLQSYWEVGFFLGASVLANDHMRVIQASEKLFKLKTPAWYLKSIVETILIYKHFVKLTTEQPVAKQELVDFWMDFLVEATKTDVTVVRFPVLILEPTKIYQPSYLSINNEVEEKTISIWHVLPDDKKGIHEWNFSASSVRGVSISKFEERCCFLYVLHNSDDFQIYFCTELHCKKFFEMVNTITEEKGRSTEEGDCESDLLEYDYEYDENGDRVVLGKGTYGIVYAGRDLSNQVRIAIKEIPERDSRYSQPLHEEIALHKHLKHKNIVQYLGSFSENGFIKIFMEQVPGGSLSALLRSKWGPLKDNEQTIGFYTKQILEGLKYLHDNQIVHRDIKGDNVLINTYSGVLKISDFGTSKRLAGINPCTETFTGTLQYMAPEIIDKGPRGYGKAADIWSLGCTIIEMATGKPPFYELGEPQAAMFKVGMFKVHPEIPESMSAEAKAFILKCFEPDPDKRACANDLLVDEFLKVSSKKKKTQPKLSALSAGSNEYLRSISLPVPVLVEDTSSSSEYGSVSPDTELKVDPFSFKTRAKSCGERDVKGIRTLFLGIPDENFEDHSAPPSPEEKDSGFFMLRKDSERRATLHRILTEDQDKIVRNLMESLAQGAEEPKLKWEHITTLIASLREFVRSTDRKIIATTLSKLKLELDFDSHGISQVQVVLFGFQDAVNKVLRNHNIKPHWMFALDSIIRKAVQTAITILVPELRPHFSLASESDTADQEDLDVEDDHEEQPSNQTVRRPQAVIEDAVATSGVSTLSSTVSHDSQSAHRSLNVQLGRMKIETNRLLEELVRKEKELQALLHRAIEEKDQEIKHLKLKSQPIEIPELPVFHLNSSGTNTEDSELTDWLRVNGADEDTISRFLAEDYTLLDVLYYVTRDDLKCLRLRGGMLCTLWKAIIDFRNKQT
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q99683 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S82 | Phosphorylation | Uniprot | |
| S83 | Phosphorylation | P11309 (PIM1) , P51812 (RPS6KA3) , P31749 (AKT1) , P31751 (AKT2) | Uniprot |
| R89 | Methylation | Uniprot | |
| S101 | Phosphorylation | Uniprot | |
| S115 | Phosphorylation | Uniprot | |
| S461 | Phosphorylation | Uniprot | |
| K465 | Ubiquitination | Uniprot | |
| T528 | Phosphorylation | Uniprot | |
| K535 | Sumoylation | Uniprot | |
| Y570 | Phosphorylation | Uniprot | |
| Y574 | Phosphorylation | Uniprot | |
| K709 | Ubiquitination | Uniprot | |
| Y718 | Phosphorylation | P23458 (JAK1) , O60674 (JAK2) | Uniprot |
| K730 | Ubiquitination | Uniprot | |
| K785 | Ubiquitination | Uniprot | |
| T813 | Phosphorylation | Q99683 (MAP3K5) | Uniprot |
| Y814 | Phosphorylation | Uniprot | |
| S821 | Phosphorylation | Uniprot | |
| T825 | Phosphorylation | Uniprot | |
| K827 | Ubiquitination | Uniprot | |
| T838 | Phosphorylation | Q14680 (MELK) , Q99683 (MAP3K5) , O95382 (MAP3K6) , Q15139 (PRKD1) | Uniprot |
| T842 | Phosphorylation | Q99683 (MAP3K5) | Uniprot |
| C869 | S-Nitrosylation | Uniprot | |
| K946 | Acetylation | Uniprot | |
| S958 | Phosphorylation | Uniprot | |
| S964 | Phosphorylation | Uniprot | |
| S966 | Phosphorylation | O15530 (PDPK1) , Q99683 (MAP3K5) | Uniprot |
| Y982 | Phosphorylation | Uniprot | |
| S986 | Phosphorylation | Uniprot | |
| S997 | Phosphorylation | Uniprot | |
| S1029 | Phosphorylation | Uniprot | |
| S1033 | Phosphorylation | Uniprot | |
| T1059 | Phosphorylation | Uniprot | |
| K1064 | Ubiquitination | Uniprot | |
| K1083 | Sumoylation | Uniprot | |
| T1109 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| K1114 | Sumoylation | Uniprot | |
| S1240 | Phosphorylation | Uniprot | |
| T1326 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
PTMs - Q99683 As Enzyme
| Substrate | Site | Source |
|---|---|---|
| O15530 (PDPK1) | S394 | Uniprot |
| O15530 (PDPK1) | S398 | Uniprot |
| P38936 (CDKN1A) | S98 | Uniprot |
| P45379-11 (TNNT2) | T191 | Uniprot |
| P45379-6 (TNNT2) | T194 | Uniprot |
| P45379-11 (TNNT2) | S195 | Uniprot |
| P45379-6 (TNNT2) | S198 | Uniprot |
| P45379-10 (TNNT2) | T201 | Uniprot |
| P45379 (TNNT2) | T204 | Uniprot |
| P45379-10 (TNNT2) | S205 | Uniprot |
| P45379 (TNNT2) | S208 | Uniprot |
| P45983 (MAPK8) | T183 | Uniprot |
| P45983 (MAPK8) | Y185 | Uniprot |
| P45985 (MAP2K4) | T261 | Uniprot |
| P46734 (MAP2K3) | S218 | Uniprot |
| P52564 (MAP2K6) | S207 | Uniprot |
| Q16539 (MAPK14) | T180 | Uniprot |
| Q16539 (MAPK14) | Y182 | Uniprot |
| Q969S3 (ZNF622) | S314 | Uniprot |
| Q969S3 (ZNF622) | T318 | Uniprot |
| Q99683 (MAP3K5) | T813 | Uniprot |
| Q99683 (MAP3K5) | T838 | Uniprot |
| Q99683 (MAP3K5) | T842 | Uniprot |
| Q99683 (MAP3K5) | S966 | Uniprot |
| Q9UER7 (DAXX) | S176 | Uniprot |
| Q9UER7 (DAXX) | S184 | Uniprot |
| Q9Y3F4 (STRAP) | T175 | Uniprot |
| Q9Y3F4 (STRAP) | S179 | Uniprot |
Research Backgrounds
Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. Plays an important role in the cascades of cellular responses evoked by changes in the environment. Mediates signaling for determination of cell fate such as differentiation and survival. Plays a crucial role in the apoptosis signal transduction pathway through mitochondria-dependent caspase activation. MAP3K5/ASK1 is required for the innate immune response, which is essential for host defense against a wide range of pathogens. Mediates signal transduction of various stressors like oxidative stress as well as by receptor-mediated inflammatory signals, such as the tumor necrosis factor (TNF) or lipopolysaccharide (LPS). Once activated, acts as an upstream activator of the MKK/JNK signal transduction cascade and the p38 MAPK signal transduction cascade through the phosphorylation and activation of several MAP kinase kinases like MAP2K4/SEK1, MAP2K3/MKK3, MAP2K6/MKK6 and MAP2K7/MKK7. These MAP2Ks in turn activate p38 MAPKs and c-jun N-terminal kinases (JNKs). Both p38 MAPK and JNKs control the transcription factors activator protein-1 (AP-1).
Phosphorylated at Thr-838 through autophosphorylation and by MAP3K6/ASK2 which leads to activation. Thr-838 is dephosphorylated by PPP5C. Ser-83 and Ser-1033 are inactivating phosphorylation sites, the former of which is phosphorylated by AKT1 and AKT2. Phosphorylated at Ser-966 which induces association of MAP3K5/ASK1 with the 14-3-3 family proteins and suppresses MAP3K5/ASK1 activity. Calcineurin (CN) dephosphorylates this site. Also dephosphorylated and activated by PGAM5.
Ubiquitinated. Tumor necrosis factor (TNF) induces TNFR2-dependent ubiquitination leading to proteasomal degradation.
Cytoplasm. Endoplasmic reticulum.
Note: Interaction with 14-3-3 proteins alters the distribution of MAP3K5/ASK1 and restricts it to the perinuclear endoplasmic reticulum region.
Abundantly expressed in heart and pancreas.
Homodimer when inactive. Binds both upstream activators and downstream substrates in multimolecular complexes. Part of a cytoplasmic complex made of HIPK1, DAB2IP and MAP3K5 in response to TNF. This complex formation promotes MAP3K5-JNK activation and subsequent apoptosis. Interacts with SOCS1 which recognizes phosphorylation of Tyr-718 and induces MAP3K5/ASK1 degradation in endothelial cells. Interacts with the 14-3-3 family proteins such as YWHAB, YWHAE, YWHAQ, YWHAH, YWHAZ and SFN. Interacts with ARRB2, BIRC2, DAB2IP, IGF1R, MAP3K6/ASK2, PGAM5, PIM1, PPP5C, SOCS1, STUB1, TRAF2, TRAF6 and TXN. Interacts with ERN1 in a TRAF2-dependent manner. Interacts with calcineurin subunit PPP3R1 and with PPM1L (By similarity). Interacts (via N-terminus) with RAF1 and this interaction inhibits the proapoptotic function of MAP3K5. Interacts with DAB2IP (via N-terminus C2 domain); the interaction occurs in a TNF-alpha-dependent manner. Interacts with DUSP13/DUSP13A; may positively regulate apoptosis. Interacts with DAXX. Interacts with RC3H2.
(Microbial infection) Interacts with HIV-1 Nef; this interaction inhibits MAP3K5 signaling.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase kinase subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Neurodegenerative diseases > Amyotrophic lateral sclerosis (ALS).
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
References
Application: WB Species: Mouse Sample:
Application: WB Species: Sample: Wild-type, Prx1KO, and Prx1OE cells
Application: WB Species: mouse Sample: liver
Application: WB Species: human Sample: LX-2 cells
Application: WB Species: rat Sample: RA cells
Application: WB Species: rat Sample: RA cells
Application: WB Species: rats Sample: Kupffer cells
Application: WB Species: Mouse Sample: BRL-3A cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.