HPSE Antibody - #DF12411
| Product: | HPSE Antibody |
| Catalog: | DF12411 |
| Description: | Rabbit polyclonal antibody to HPSE |
| Application: | WB IHC |
| Reactivity: | Human, Mouse |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 60 kDa; 61kD(Calculated). |
| Uniprot: | Q9Y251 |
| RRID: | AB_2845216 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12411, RRID:AB_2845216.
Fold/Unfold
Endo glucoronidase; Endo-glucoronidase; HEP; Heparanase 50 kDa subunit; Heparanase; Heparanase-1; Heparanase1; Hpa 1; HPA; Hpa1; HPR 1; HPR1; HPSE 1; HPSE; HPSE_HUMAN; HPSE1; HSE 1; HSE1;
Immunogens
Highly expressed in placenta and spleen and weakly expressed in lymph node, thymus, peripheral blood leukocytes, bone marrow, endothelial cells, fetal liver and tumor tissues. Also expressed in hair follicles, specifically in both Henle's and Huxley's layers of inner the root sheath (IRS) at anagen phase.
- Q9Y251 HPSE_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MLLRSKPALPPPLMLLLLGPLGPLSPGALPRPAQAQDVVDLDFFTQEPLHLVSPSFLSVTIDANLATDPRFLILLGSPKLRTLARGLSPAYLRFGGTKTDFLIFDPKKESTFEERSYWQSQVNQDICKYGSIPPDVEEKLRLEWPYQEQLLLREHYQKKFKNSTYSRSSVDVLYTFANCSGLDLIFGLNALLRTADLQWNSSNAQLLLDYCSSKGYNISWELGNEPNSFLKKADIFINGSQLGEDFIQLHKLLRKSTFKNAKLYGPDVGQPRRKTAKMLKSFLKAGGEVIDSVTWHHYYLNGRTATKEDFLNPDVLDIFISSVQKVFQVVESTRPGKKVWLGETSSAYGGGAPLLSDTFAAGFMWLDKLGLSARMGIEVVMRQVFFGAGNYHLVDENFDPLPDYWLSLLFKKLVGTKVLMASVQGSKRRKLRVYLHCTNTDNPRYKEGDLTLYAINLHNVTKYLRLPYPFSNKQVDKYLLRPLGPHGLLSKSVQLNGLTLKMVDDQTLPPLMEKPLRPGSSLGLPAFSYSFFVIRNAKVAACI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q9Y251 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K98 | Acetylation | Uniprot | |
| T99 | Phosphorylation | Uniprot | |
| Y156 | Phosphorylation | Uniprot | |
| N162 | N-Glycosylation | Uniprot | |
| Y165 | Phosphorylation | Uniprot | |
| N178 | N-Glycosylation | Uniprot | |
| N200 | N-Glycosylation | Uniprot | |
| N217 | N-Glycosylation | Uniprot | |
| N238 | N-Glycosylation | Uniprot | |
| K262 | Ubiquitination | Uniprot | |
| S332 | Phosphorylation | Uniprot | |
| T333 | Phosphorylation | Uniprot | |
| Y348 | Phosphorylation | Uniprot | |
| K427 | Acetylation | Uniprot | |
| N459 | N-Glycosylation | Uniprot | |
| T507 | Phosphorylation | Uniprot |
Research Backgrounds
Endoglycosidase that cleaves heparan sulfate proteoglycans (HSPGs) into heparan sulfate side chains and core proteoglycans. Participates in extracellular matrix (ECM) degradation and remodeling. Selectively cleaves the linkage between a glucuronic acid unit and an N-sulfo glucosamine unit carrying either a 3-O-sulfo or a 6-O-sulfo group. Can also cleave the linkage between a glucuronic acid unit and an N-sulfo glucosamine unit carrying a 2-O-sulfo group, but not linkages between a glucuronic acid unit and a 2-O-sulfated iduronic acid moiety. It is essentially inactive at neutral pH but becomes active under acidic conditions such as during tumor invasion and in inflammatory processes. Facilitates cell migration associated with metastasis, wound healing and inflammation. Enhances shedding of syndecans, and increases endothelial invasion and angiogenesis in myelomas. Acts as procoagulant by increasing the generation of activation factor X in the presence of tissue factor and activation factor VII. Increases cell adhesion to the extracellular matrix (ECM), independent of its enzymatic activity. Induces AKT1/PKB phosphorylation via lipid rafts increasing cell mobility and invasion. Heparin increases this AKT1/PKB activation. Regulates osteogenesis. Enhances angiogenesis through up-regulation of SRC-mediated activation of VEGF. Implicated in hair follicle inner root sheath differentiation and hair homeostasis.
Proteolytically processed. The cleavage of the 65 kDa form leads to the generation of a linker peptide, and 8 kDa and 50 kDa products. The active form, the 8/50 kDa heterodimer, is resistant to degradation. Complete removal of the linker peptide appears to be a prerequisite to the complete activation of the enzyme.
N-glycosylated. Glycosylation of the 50 kDa subunit appears to be essential for its solubility.
Lysosome membrane>Peripheral membrane protein. Secreted. Nucleus.
Note: Proheparanase is secreted via vesicles of the Golgi. Interacts with cell membrane heparan sulfate proteoglycans (HSPGs). Endocytosed and accumulates in endosomes. Transferred to lysosomes where it is proteolytically cleaved to produce the active enzyme. Under certain stimuli, transferred to the cell surface. Associates with lipid rafts. Colocalizes with SDC1 in endosomal/lysosomal vesicles. Accumulates in perinuclear lysosomal vesicles. Heparin retains proheparanase in the extracellular medium (By similarity).
Highly expressed in placenta and spleen and weakly expressed in lymph node, thymus, peripheral blood leukocytes, bone marrow, endothelial cells, fetal liver and tumor tissues. Also expressed in hair follicles, specifically in both Henle's and Huxley's layers of inner the root sheath (IRS) at anagen phase.
Heterodimer; heterodimer formation between the 8 kDa and the 50 kDa subunits is required for enzyme activity. Interacts with TF; the interaction, inhibited by heparin, enhances the generation of activated factor X and activates coagulation. Interacts with HRG; the interaction is enhanced at acidic pH, partially inhibits binding of HPSE to cell surface receptors and modulates its enzymatic activity. Interacts with SDC1; the interaction enhances the shedding of SDC1. Interacts with HPSE2.
Belongs to the glycosyl hydrolase 79 family.
Research Fields
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Metabolism > Glycan biosynthesis and metabolism > Glycosaminoglycan degradation.
· Metabolism > Global and overview maps > Metabolic pathways.
References
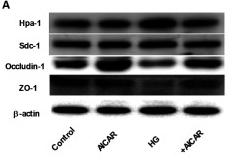
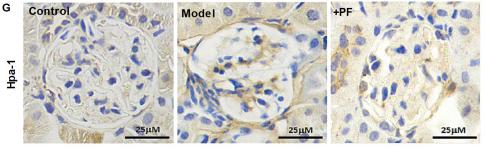
Application: WB Species: Mouse Sample: lung tissue
Application: WB Species: Human Sample: glomerular endothelial cells (GEnCs)
Application: IHC Species: Human Sample: glomerular endothelial cells (GEnCs)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.