Phospho-PTEN (Ser370) Antibody - #AF3351
| Product: | Phospho-PTEN (Ser370) Antibody |
| Catalog: | AF3351 |
| Description: | Rabbit polyclonal antibody to Phospho-PTEN (Ser370) |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 54kDa; 47kD(Calculated). |
| Uniprot: | P60484 |
| RRID: | AB_2834766 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3351, RRID:AB_2834766.
Fold/Unfold
10q23del; BZS; DEC; GLM2; MGC11227; MHAM; MMAC1; MMAC1 phosphatase and tensin homolog deleted on chromosome 10; Mutated in multiple advanced cancers 1; Phosphatase and tensin homolog; Phosphatase and tensin like protein; Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN; Pten; PTEN_HUMAN; PTEN1; TEP1;
Immunogens
Expressed at a relatively high level in all adult tissues, including heart, brain, placenta, lung, liver, muscle, kidney and pancreas.
- P60484 PTEN_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTAIIKEIVSRNKRRYQEDGFDLDLTYIYPNIIAMGFPAERLEGVYRNNIDDVVRFLDSKHKNHYKIYNLCAERHYDTAKFNCRVAQYPFEDHNPPQLELIKPFCEDLDQWLSEDDNHVAAIHCKAGKGRTGVMICAYLLHRGKFLKAQEALDFYGEVRTRDKKGVTIPSQRRYVYYYSYLLKNHLDYRPVALLFHKMMFETIPMFSGGTCNPQFVVCQLKVKIYSSNSGPTRREDKFMYFEFPQPLPVCGDIKVEFFHKQNKMLKKDKMFHFWVNTFFIPGPEETSEKVENGSLCDQEIDSICSIERADNDKEYLVLTLTKNDLDKANKDKANRYFSPNFKVKLYFTKTVEEPSNPEASSSTSVTPDVSDNEPDHYRYSDTTDSDPENEPFDEDQHTQITKV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P60484 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Ubiquitination | Uniprot | ||
| T2 | Acetylation | Uniprot | |
| K6 | Acetylation | Uniprot | |
| K6 | Ubiquitination | Uniprot | |
| K13 | Ubiquitination | Uniprot | |
| Y27 | Phosphorylation | P06213 (INSR) | Uniprot |
| Y46 | Phosphorylation | P12931 (SRC) | Uniprot |
| K66 | Ubiquitination | Uniprot | |
| Y68 | Phosphorylation | P12931 (SRC) | Uniprot |
| K80 | Ubiquitination | Uniprot | |
| S113 | Phosphorylation | Q13315 (ATM) | Uniprot |
| K125 | Acetylation | Uniprot | |
| K128 | Acetylation | Uniprot | |
| Y138 | Phosphorylation | Uniprot | |
| Y155 | Phosphorylation | P12931 (SRC) | Uniprot |
| K163 | Acetylation | Uniprot | |
| K164 | Acetylation | Uniprot | |
| Y174 | Phosphorylation | P12931 (SRC) , P06213 (INSR) | Uniprot |
| Y176 | Phosphorylation | Uniprot | |
| Y177 | Phosphorylation | Uniprot | |
| Y178 | Phosphorylation | Uniprot | |
| S179 | Phosphorylation | Uniprot | |
| Y180 | Phosphorylation | Uniprot | |
| K223 | Ubiquitination | Uniprot | |
| S227 | Phosphorylation | Uniprot | |
| S229 | Phosphorylation | Q13464 (ROCK1) | Uniprot |
| T232 | Phosphorylation | Q13464 (ROCK1) | Uniprot |
| Y240 | Phosphorylation | P12931 (SRC) , P21802 (FGFR2) , P07948 (LYN) , P22607 (FGFR3) , P06239 (LCK) | Uniprot |
| K254 | Sumoylation | Uniprot | |
| K266 | Sumoylation | Uniprot | |
| K289 | Sumoylation | Uniprot | |
| K289 | Ubiquitination | Uniprot | |
| S294 | Phosphorylation | Uniprot | |
| S302 | Phosphorylation | Uniprot | |
| Y315 | Phosphorylation | P12931 (SRC) , P06239 (LCK) | Uniprot |
| T319 | Phosphorylation | Uniprot | |
| T321 | Phosphorylation | Uniprot | |
| K332 | Acetylation | Uniprot | |
| Y336 | Phosphorylation | P42685 (FRK) | Uniprot |
| S362 | Phosphorylation | P49841 (GSK3B) | Uniprot |
| T366 | Phosphorylation | P49841 (GSK3B) , Q9H4B4 (PLK3) | Uniprot |
| S370 | Phosphorylation | P68400 (CSNK2A1) , Q9H4B4 (PLK3) | Uniprot |
| Y377 | Phosphorylation | P12931 (SRC) | Uniprot |
| S380 | Phosphorylation | Q15831 (STK11) , Q05513 (PRKCZ) , P68400 (CSNK2A1) , P53350 (PLK1) | Uniprot |
| T382 | Phosphorylation | Q15831 (STK11) , P68400 (CSNK2A1) , Q05513 (PRKCZ) , P53350 (PLK1) | Uniprot |
| T383 | Phosphorylation | P53350 (PLK1) , P68400 (CSNK2A1) , Q05513 (PRKCZ) , Q15831 (STK11) | Uniprot |
| S385 | Phosphorylation | Q15831 (STK11) , P68400 (CSNK2A1) | Uniprot |
| T398 | Phosphorylation | Q13315 (ATM) | Uniprot |
| T401 | Phosphorylation | Uniprot | |
| K402 | Acetylation | Uniprot |
Research Backgrounds
Tumor suppressor. Acts as a dual-specificity protein phosphatase, dephosphorylating tyrosine-, serine- and threonine-phosphorylated proteins. Also acts as a lipid phosphatase, removing the phosphate in the D3 position of the inositol ring from phosphatidylinositol 3,4,5-trisphosphate, phosphatidylinositol 3,4-diphosphate, phosphatidylinositol 3-phosphate and inositol 1,3,4,5-tetrakisphosphate with order of substrate preference in vitro PtdIns(3,4,5)P3 > PtdIns(3,4)P2 > PtdIns3P > Ins(1,3,4,5)P4. The lipid phosphatase activity is critical for its tumor suppressor function. Antagonizes the PI3K-AKT/PKB signaling pathway by dephosphorylating phosphoinositides and thereby modulating cell cycle progression and cell survival. The unphosphorylated form cooperates with AIP1 to suppress AKT1 activation. Dephosphorylates tyrosine-phosphorylated focal adhesion kinase and inhibits cell migration and integrin-mediated cell spreading and focal adhesion formation. Plays a role as a key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development and synapse formation. May be a negative regulator of insulin signaling and glucose metabolism in adipose tissue. The nuclear monoubiquitinated form possesses greater apoptotic potential, whereas the cytoplasmic nonubiquitinated form induces less tumor suppressive ability. In motile cells, suppresses the formation of lateral pseudopods and thereby promotes cell polarization and directed movement.
Functional kinase, like isoform 1 it antagonizes the PI3K-AKT/PKB signaling pathway. Plays a role in mitochondrial energetic metabolism by promoting COX activity and ATP production, via collaboration with isoform 1 in increasing protein levels of PINK1.
Constitutively phosphorylated by CK2 under normal conditions. Phosphorylated in vitro by MAST1, MAST2, MAST3 and STK11. Phosphorylation results in an inhibited activity towards PIP3. Phosphorylation can both inhibit or promote PDZ-binding. Phosphorylation at Tyr-336 by FRK/PTK5 protects this protein from ubiquitin-mediated degradation probably by inhibiting its binding to NEDD4. Phosphorylation by ROCK1 is essential for its stability and activity. Phosphorylation by PLK3 promotes its stability and prevents its degradation by the proteasome.
Monoubiquitinated; monoubiquitination is increased in presence of retinoic acid. Deubiquitinated by USP7; leading to its nuclear exclusion. Monoubiquitination of one of either Lys-13 and Lys-289 amino acid is sufficient to modulate PTEN compartmentalization. Ubiquitinated by XIAP/BIRC4.
Cytoplasm. Nucleus. Nucleus>PML body.
Note: Monoubiquitinated form is nuclear. Nonubiquitinated form is cytoplasmic. Colocalized with PML and USP7 in PML nuclear bodies (PubMed:18716620). XIAP/BIRC4 promotes its nuclear localization (PubMed:19473982).
Secreted.
Note: May be secreted via a classical signal peptide and reenter into cells with the help of a poly-Arg motif.
Expressed at a relatively high level in all adult tissues, including heart, brain, placenta, lung, liver, muscle, kidney and pancreas.
Monomer. The unphosphorylated form interacts with the second PDZ domain of AIP1 and with DLG1 and MAST2 in vitro. Interacts with MAGI2, MAGI3, MAST1 and MAST3, but neither with MAST4 nor with DLG5; interaction with MAGI2 increases protein stability. Interacts with NEDD4. Interacts with NDFIP1 and NDFIP2; in the presence of NEDD4 or ITCH, this interaction promotes PTEN ubiquitination. Interacts (via C2 domain) with FRK. Interacts with USP7; the interaction is direct. Interacts with ROCK1 (By similarity). Interacts with XIAP/BIRC4. Interacts with STK11; the interaction phosphorylates PTEN. Interacts with PPP1R16B. Interacts with NOP53; regulates PTEN phosphorylation and increases its stability.
The C2 domain binds phospholipid membranes in vitro in a Ca(2+)-independent manner; this binding is important for its tumor suppressor function.
Belongs to the PTEN phosphatase protein family.
Research Fields
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phosphatidylinositol signaling system.
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Metabolism > Carbohydrate metabolism > Inositol phosphate metabolism.
References
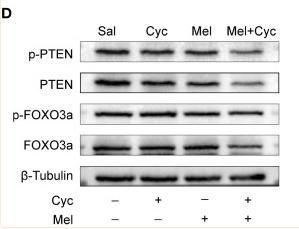
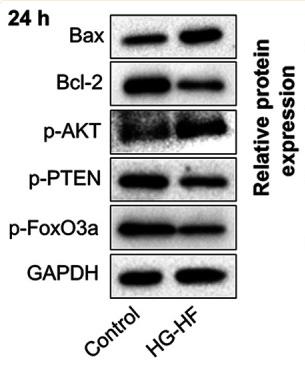
Application: WB Species: Mice Sample:
Application: WB Species: Human Sample: PHEGFs
Application: WB Species: Rat Sample: H9c2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.