Phospho-RIP3 (Ser232) Antibody - #AF7443
| Product: | Phospho-RIP3 (Ser232) Antibody |
| Catalog: | AF7443 |
| Description: | Rabbit polyclonal antibody to Phospho-RIP3 (Ser232) |
| Application: | WB |
| Reactivity: | Mouse, Rat |
| Mol.Wt.: | 57kDa.; 53kD(Calculated). |
| Uniprot: | Q9QZL0 |
| RRID: | AB_2843883 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7443, RRID:AB_2843883.
Fold/Unfold
Receptor interacting protein 3; Receptor interacting serine threonine kinase 3; Receptor interacting serine/threonine protein kinase 3; Receptor-interacting protein 3; Receptor-interacting serine/threonine-protein kinase 3; RIP 3; RIP like protein kinase 3; RIP-3; RIP-like protein kinase 3; RIPK 3; RIPK3; RIPK3_HUMAN;
Immunogens
A synthesized peptide derived from mouse RIP3 around the phosphorylation site of Ser232.
- Q9QZL0 RIPK3_MOUSE:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSSVKLWPTGASAVPLVSREELKKLEFVGKGGFGVVFRAHHRTWNHDVAVKIVNSKKISWEVKAMVNLRNENVLLLLGVTEDLQWDFVSGQALVTRFMENGSLAGLLQPECPRPWPLLCRLLQEVVLGMCYLHSLNPPLLHRDLKPSNILLDPELHAKLADFGLSTFQGGSQSGSGSGSGSRDSGGTLAYLDPELLFDVNLKASKASDVYSFGILVWAVLAGREAELVDKTSLIRETVCDRQSRPPLTELPPGSPETPGLEKLKELMIHCWGSQSENRPSFQDCEPKTNEVYNLVKDKVDAAVSEVKHYLSQHRSSGRNLSAREPSQRGTEMDCPRETMVSKMLDRLHLEEPSGPVPGKCPERQAQDTSVGPATPARTSSDPVAGTPQIPHTLPFRGTTPGPVFTETPGPHPQRNQGDGRHGTPWYPWTPPNPMTGPPALVFNNCSEVQIGNYNSLVAPPRTTASSSAKYDQAQFGRGRGWQPFHK
PTMs - Q9QZL0 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S2 | Phosphorylation | Uniprot | |
| K5 | Ubiquitination | Uniprot | |
| S165 | Phosphorylation | Uniprot | |
| S184 | Phosphorylation | Uniprot | |
| T231 | Phosphorylation | Uniprot | |
| S232 | Phosphorylation | Q9QZL0 (Ripk3) | Uniprot |
| T257 | Phosphorylation | Uniprot | |
| K264 | Ubiquitination | Uniprot | |
| S304 | Phosphorylation | Uniprot | |
| S326 | Phosphorylation | Uniprot | |
| T338 | Phosphorylation | Uniprot | |
| S353 | Phosphorylation | Uniprot | |
| S369 | Phosphorylation | Uniprot | |
| T374 | Phosphorylation | Uniprot | |
| S380 | Phosphorylation | Uniprot | |
| T392 | Phosphorylation | Uniprot | |
| T399 | Phosphorylation | P63085 (Mapk1) | Uniprot |
| R477 | Methylation | Uniprot | |
| R479 | Methylation | Uniprot |
PTMs - Q9QZL0 As Enzyme
Research Backgrounds
Serine/threonine-protein kinase that activates necroptosis and apoptosis, two parallel forms of cell death. Necroptosis, a programmed cell death process in response to death-inducing TNF-alpha family members, is triggered by RIPK3 following activation by ZBP1. Activated RIPK3 forms a necrosis-inducing complex and mediates phosphorylation of MLKL, promoting MLKL localization to the plasma membrane and execution of programmed necrosis characterized by calcium influx and plasma membrane damage. In addition to TNF-induced necroptosis, necroptosis can also take place in the nucleus in response to orthomyxoviruses infection: following ZBP1 activation, which senses double-stranded Z-RNA structures, nuclear RIPK3 catalyzes phosphorylation and activation of MLKL, promoting disruption of the nuclear envelope and leakage of cellular DNA into the cytosol. Also regulates apoptosis: apoptosis depends on RIPK1, FADD and CASP8, and is independent of MLKL and RIPK3 kinase activity. Phosphorylates RIPK1: RIPK1 and RIPK3 undergo reciprocal auto- and trans-phosphorylation (By similarity). In some cell types, also able to restrict viral replication by promoting cell death-independent responses. In response to flavivirus infection in neurons, promotes a cell death-independent pathway that restricts viral replication: together with ZBP1, promotes a death-independent transcriptional program that modifies the cellular metabolism via up-regulation expression of the enzyme ACOD1/IRG1 and production of the metabolite itaconate. Itaconate inhibits the activity of succinate dehydrogenase, generating a metabolic state in neurons that suppresses replication of viral genomes. RIPK3 binds to and enhances the activity of three metabolic enzymes: GLUL, GLUD1, and PYGL (By similarity). These metabolic enzymes may eventually stimulate the tricarboxylic acid cycle and oxidative phosphorylation, which could result in enhanced ROS production (By similarity).
RIPK1 and RIPK3 undergo reciprocal auto- and trans-phosphorylation (By similarity). Autophosphorylated following interaction with ZBP1. Phosphorylation of Ser-204 plays a role in the necroptotic function of RIPK3 (By similarity). Autophosphorylates at Thr-231 and Ser-232 following activation by ZBP1: phosphorylation at these sites is a hallmark of necroptosis and is required for binding MLKL. Phosphorylation at Thr-187 is important for its kinase activity, interaction with PELI1 and for its ability to mediate TNF-induced necroptosis (By similarity).
Polyubiquitinated with 'Lys-48' and 'Lys-63'-linked chains by BIRC2/c-IAP1 and BIRC3/c-IAP2, leading to activation of NF-kappa-B. Ubiquitinated by STUB1 leading to its subsequent proteasome-dependent degradation.
Cytoplasm>Cytosol. Nucleus.
Note: Mainly cytoplasmic (PubMed:32200799, PubMed:32296175). Present in the nucleus in response to influenza A virus (IAV) infection (PubMed:32200799).
Expressed in embryo and in adult spleen, liver, testis, heart, brain and lung.
Interacts (via RIP homotypic interaction motif) with RIPK1 (via RIP homotypic interaction motif); this interaction induces RIPK1 phosphorylation and formation of a RIPK1-RIPK3 necrosis-inducing complex. Interacts with MLKL; the interaction is direct and triggers necroptosis. Interacts with ZBP1 (via RIP homotypic interaction motif); interaction with ZBP1 activates RIPK3, triggering necroptosis. Upon TNF-induced necrosis, the RIPK1-RIPK3 dimer further interacts with PGAM5 and MLKL; the formation of this complex leads to PGAM5 phosphorylation and increase in PGAM5 phosphatase activity (By similarity). Binds TRAF2 and is recruited to the TNFR-1 signaling complex (By similarity). Interacts with PYGL, GLUL and GLUD1; these interactions result in activation of these metabolic enzymes (By similarity). Interacts with BIRC2/c-IAP1, BIRC3/c-IAP2 and XIAP/BIRC4 (By similarity). Interacts with ARHGEF2 (By similarity). Interacts with PELI1 (via atypical FHA domain); the phosphorylated form at Thr-187 binds preferentially to PELI1. Interacts with BUB1B, TRAF2 and STUB1 (By similarity). Interacts with CASP6 (By similarity).
(Microbial infection) Interacts (via RIP homotypic interaction motif) with murid herpesvirus protein RIR1; this interaction disrupts RIP3-RIP1 interactions characteristic of TNF-alpha induced necroptosis, thereby suppressing this death pathway.
The RIP homotypic interaction motif (RHIM) mediates interaction with the RHIM motif of RIPK1. Both motifs form a hetero-amyloid serpentine fold, stabilized by hydrophobic packing and featuring an unusual Cys-Ser ladder of alternating Ser (from RIPK1) and Cys (from RIPK3).
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family.
References
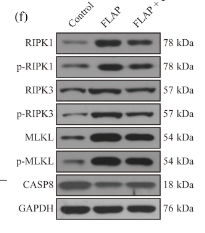
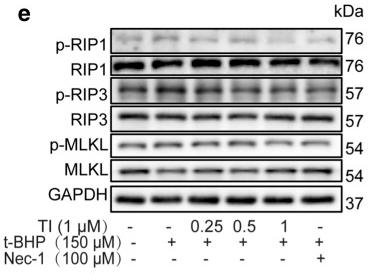
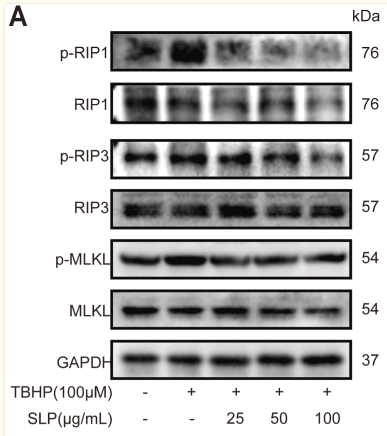
Application: WB Species: Rat Sample: apoptotic and necrotic cells
Application: WB Species: Mouse Sample: skin tissues
Application: WB Species: mouse Sample: myocardium
Application: WB Species: Rat Sample: H9c2 cells
Application: WB Species: Rat Sample:
Application: WB Species: Rat Sample: H9c2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.