Phospho-PKA alpha/beta/gamma CAT (Thr198) Antibody - #AF7246
| Product: | Phospho-PKA alpha/beta/gamma CAT (Thr198) Antibody |
| Catalog: | AF7246 |
| Description: | Rabbit polyclonal antibody to Phospho-PKA alpha/beta/gamma CAT (Thr198) |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Dog, Xenopus |
| Mol.Wt.: | 40kDa; 41kD,40kD(Calculated). |
| Uniprot: | P17612 | P22694 | P22612 |
| RRID: | AB_2843686 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7246, RRID:AB_2843686.
Fold/Unfold
cAMP dependent protein kinase alpha catalytic subunit; cAMP dependent protein kinase beta catalytic subunit; cAMP dependent protein kinase catalytic beta subunit isoform 4ab; cAMP dependent protein kinase catalytic subunit alpha; cAMP dependent protein kinase catalytic subunit alpha, isoform 1; cAMP dependent protein kinase catalytic subunit beta; cAMP-dependent protein kinase catalytic subunit alpha; KAPCA_HUMAN; PKA C alpha; PKA C beta; PKA C-alpha; PKACA; PKACB; PPNAD4; PRKACA; PRKACAA; PRKACB; Protein kinase A catalytic subunit alpha; Protein kinase A catalytic subunit; Protein kinase A catalytic subunit beta; Protein kinase, cAMP dependent, catalytic, alpha; Protein kinase, cAMP dependent, catalytic, beta; cAMP-dependent protein kinase catalytic beta subunit isoform 4ab; cAMP-dependent protein kinase catalytic subunit beta; KAPCB_HUMAN; PKA C beta; PKA C-beta; PKACB; Prkacb; protein kinase A catalytic subunit beta; Protein kinase cAMP dependent catalytic beta; cAMP-dependent protein kinase catalytic subunit gamma; KAPCG_HUMAN; KAPG; PKA C gamma; PKA C-gamma; PKACg; PRKACG; Protein kinase cAMP dependent catalytic gamma; Serine (threonine) protein kinase;
Immunogens
Isoform 1 is ubiquitous. Isoform 2 is sperm-specific and is enriched in pachytene spermatocytes but is not detected in round spermatids.
P22694 KAPCB_HUMAN:Isoform 1 is most abundant in the brain, with low level expression in kidney. Isoform 2 is predominantly expressed in thymus, spleen and kidney. Isoform 3 and isoform 4 are only expressed in the brain.
P22612 KAPCG_HUMAN:Testis specific. But important tissues such as brain and ovary have not been analyzed for the content of transcript.
- P17612 KAPCA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
- P22694 KAPCB_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGNAATAKKGSEVESVKEFLAKAKEDFLKKWENPTQNNAGLEDFERKKTLGTGSFGRVMLVKHKATEQYYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVRLEYAFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDHQGYIQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVSDIKTHKWFATTDWIAIYQRKVEAPFIPKFRGSGDTSNFDDYEEEDIRVSITEKCAKEFGEF
- P22612 KAPCG_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGNAPAKKDTEQEESVNEFLAKARGDFLYRWGNPAQNTASSDQFERLRTLGMGSFGRVMLVRHQETGGHYAMKILNKQKVVKMKQVEHILNEKRILQAIDFPFLVKLQFSFKDNSYLYLVMEYVPGGEMFSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAVGFPPFYADQPIQIYEKIVSGRVRFPSKLSSDLKHLLRSLLQVDLTKRFGNLRNGVGDIKNHKWFATTSWIAIYEKKVEAPFIPKYTGPGDASNFDDYEEEELRISINEKCAKEFSEF
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P17612/P22694/P22612 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| G2 | Myristoylation | Uniprot | |
| S11 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| S15 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K17 | Ubiquitination | Uniprot | |
| K24 | Ubiquitination | Uniprot | |
| K30 | Ubiquitination | Uniprot | |
| K48 | Ubiquitination | Uniprot | |
| T49 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| T52 | Phosphorylation | Uniprot | |
| S54 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| Y70 | Phosphorylation | Uniprot | |
| K84 | Ubiquitination | Uniprot | |
| T89 | Phosphorylation | Uniprot | |
| K93 | Ubiquitination | Uniprot | |
| S140 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K190 | Ubiquitination | Uniprot | |
| T196 | Phosphorylation | Uniprot | |
| T198 | Phosphorylation | O15530 (PDPK1) , P17612 (PRKACA) | Uniprot |
| T202 | Phosphorylation | Uniprot | |
| S213 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K214 | Ubiquitination | Uniprot | |
| K255 | Ubiquitination | Uniprot | |
| S260 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| S264 | Phosphorylation | Uniprot | |
| K267 | Acetylation | Uniprot | |
| K267 | Ubiquitination | Uniprot | |
| K280 | Acetylation | Uniprot | |
| K280 | Ubiquitination | Uniprot | |
| K286 | Ubiquitination | Uniprot | |
| K293 | Ubiquitination | Uniprot | |
| K310 | Ubiquitination | Uniprot | |
| K318 | Ubiquitination | Uniprot | |
| K320 | Ubiquitination | Uniprot | |
| S326 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| Y331 | Phosphorylation | Uniprot | |
| S339 | Phosphorylation | P17612 (PRKACA) | Uniprot |
| K343 | Ubiquitination | Uniprot | |
| K346 | Ubiquitination | Uniprot |
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| G2 | Myristoylation | Uniprot | |
| T6 | Phosphorylation | Uniprot | |
| S11 | Phosphorylation | Uniprot | |
| S15 | Phosphorylation | Uniprot | |
| K17 | Ubiquitination | Uniprot | |
| K24 | Ubiquitination | Uniprot | |
| K30 | Ubiquitination | Uniprot | |
| K47 | Acetylation | Uniprot | |
| K48 | Ubiquitination | Uniprot | |
| T49 | Phosphorylation | Uniprot | |
| T52 | Phosphorylation | Uniprot | |
| S54 | Phosphorylation | Uniprot | |
| K64 | Ubiquitination | Uniprot | |
| Y69 | Phosphorylation | Uniprot | |
| Y70 | Phosphorylation | Uniprot | |
| K73 | Ubiquitination | Uniprot | |
| K84 | Ubiquitination | Uniprot | |
| T89 | Phosphorylation | Uniprot | |
| K93 | Ubiquitination | Uniprot | |
| Y157 | Phosphorylation | Uniprot | |
| Y165 | Phosphorylation | Uniprot | |
| Y180 | Phosphorylation | Uniprot | |
| T184 | Phosphorylation | Uniprot | |
| T196 | Phosphorylation | Uniprot | |
| T198 | Phosphorylation | Uniprot | |
| T202 | Phosphorylation | Uniprot | |
| K214 | Ubiquitination | Uniprot | |
| K255 | Ubiquitination | Uniprot | |
| S260 | Phosphorylation | Uniprot | |
| S264 | Phosphorylation | Uniprot | |
| K267 | Acetylation | Uniprot | |
| K267 | Ubiquitination | Uniprot | |
| K280 | Acetylation | Uniprot | |
| K280 | Ubiquitination | Uniprot | |
| K286 | Ubiquitination | Uniprot | |
| K293 | Ubiquitination | Uniprot | |
| K310 | Ubiquitination | Uniprot | |
| K318 | Ubiquitination | Uniprot | |
| S322 | Phosphorylation | Uniprot | |
| T325 | Phosphorylation | Uniprot | |
| S326 | Phosphorylation | Uniprot | |
| Y331 | Phosphorylation | Uniprot | |
| S339 | Phosphorylation | P22694 (PRKACB) | Uniprot |
| T341 | Phosphorylation | Uniprot | |
| K343 | Ubiquitination | Uniprot |
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Y118 | Phosphorylation | Uniprot | |
| Y123 | Phosphorylation | Uniprot | |
| R132 | Methylation | Uniprot | |
| T196 | Phosphorylation | Uniprot | |
| T198 | Phosphorylation | Uniprot | |
| T202 | Phosphorylation | Uniprot | |
| K214 | Ubiquitination | Uniprot | |
| K310 | Ubiquitination | Uniprot |
PTMs - P17612/P22694/P22612 As Enzyme
| Substrate | Site | Source |
|---|---|---|
| O00141 (SGK1) | T369 | Uniprot |
| O00168 (FXYD1) | S83 | Uniprot |
| O00168 (FXYD1) | S88 | Uniprot |
| O00231 (PSMD11) | S14 | Uniprot |
| O00418 (EEF2K) | S366 | Uniprot |
| O00418 (EEF2K) | S500 | Uniprot |
| O00429 (DNM1L) | S637 | Uniprot |
| O00482 (NR5A2) | S510 | Uniprot |
| O14558 (HSPB6) | S16 | Uniprot |
| O14649 (KCNK3) | S393 | Uniprot |
| O14813 (PHOX2A) | S153 | Uniprot |
| O14921 (RGS13) | T41 | Uniprot |
| O14936 (CASK) | S562 | Uniprot |
| O14936 (CASK) | T741 | Uniprot |
| O14965 (AURKA) | T288 | Uniprot |
| O14974 (PPP1R12A) | S695 | Uniprot |
| O14974 (PPP1R12A) | T696 | Uniprot |
| O14974 (PPP1R12A) | S852 | Uniprot |
| O14974 (PPP1R12A) | T853 | Uniprot |
| O15217 (GSTA4) | S189 | Uniprot |
| O15392 (BIRC5) | S20 | Uniprot |
| O15551 (CLDN3) | T192 | Uniprot |
| O15554 (KCNN4) | S334 | Uniprot |
| O43164 (PJA2) | S342 | Uniprot |
| O43164 (PJA2) | T389 | Uniprot |
| O43181 (NDUFS4) | S173 | Uniprot |
| O43566-7 (RGS14) | S260 | Uniprot |
| O43566-7 (RGS14) | T495 | Uniprot |
| O43602 (DCX) | S128 | Uniprot |
| O43665-2 (RGS10) | S162 | Uniprot |
| O43665-3 (RGS10) | S176 | Uniprot |
| O60240 (PLIN1) | S81 | Uniprot |
| O60479 (DLX3) | S10 | Uniprot |
| O60716 (CTNND1) | S879 | Uniprot |
| O60825-1 (PFKFB2) | S466 | Uniprot |
| O60928 (KCNJ13) | S287 | Uniprot |
| O60941 (DTNB) | T11 | Uniprot |
| O60941 (DTNB) | T424 | Uniprot |
| O75376 (NCOR1) | S70 | Uniprot |
| O75581 (LRP6) | T1548 | Uniprot |
| O75688 (PPM1B) | S195 | Uniprot |
| O75899 (GABBR2) | S893 | Uniprot |
| O76054 (SEC14L2) | S289 | Uniprot |
| O76074 (PDE5A) | S102 | Uniprot |
| O94811 (TPPP) | S32 | Uniprot |
| O94811 (TPPP) | T92 | Uniprot |
| O94811 (TPPP) | S159 | Uniprot |
| O95180 (CACNA1H) | S1107 | Uniprot |
| O95180 (CACNA1H) | S1144 | Uniprot |
| O95295 (SNAPIN) | S50 | Uniprot |
| O95477 (ABCA1) | S1042 | Uniprot |
| O95477 (ABCA1) | S2054 | Uniprot |
| O95644 (NFATC1) | S245 | Uniprot |
| O95644 (NFATC1) | S269 | Uniprot |
| O95644 (NFATC1) | S294 | Uniprot |
| O95863 (SNAI1) | S11 | Uniprot |
| O96001-1 (PPP1R17) | T68 | Uniprot |
| O96001-1 (PPP1R17) | T119 | Uniprot |
| O96004 (HAND1) | S98 | Uniprot |
| O96004 (HAND1) | T107 | Uniprot |
| O96004 (HAND1) | S109 | Uniprot |
| P00439 (PAH) | S16 | Uniprot |
| P01100-1 (FOS) | S362 | Uniprot |
| P01892 (HLA-A) | S337 | Uniprot |
| P01892 (HLA-A) | S343 | Uniprot |
| P02686-6 (MBP) | S8 | Uniprot |
| P02686-4 (MBP) | S13 | Uniprot |
| P02686-6 (MBP) | T36 | Uniprot |
| P02686-3 (MBP) | S57 | Uniprot |
| P02686-6 (MBP) | S122 | Uniprot |
| P02686-5 (MBP) | S133 | Uniprot |
| P02686-2 (MBP) | S141 | Uniprot |
| P02686 (MBP) | S146 | Uniprot |
| P02686-4 (MBP) | S148 | Uniprot |
| P02686-6 (MBP) | S151 | Uniprot |
| P02686-3 (MBP) | S159 | Uniprot |
| P02686-5 (MBP) | S162 | Uniprot |
| P02686 (MBP) | T169 | Uniprot |
| P02686-4 (MBP) | S177 | Uniprot |
| P02686-3 (MBP) | S188 | Uniprot |
| P02686 (MBP) | S190 | Uniprot |
| P02686 (MBP) | S249 | Uniprot |
| P02686 (MBP) | S266 | Uniprot |
| P02686 (MBP) | S295 | Uniprot |
| P03372 (ESR1) | S236 | Uniprot |
| P03372 (ESR1) | S305 | Uniprot |
| P04004 (VTN) | S397 | Uniprot |
| P04035-1 (HMGCR) | S872 | Uniprot |
| P04049-1 (RAF1) | S43 | Uniprot |
| P04049-1 (RAF1) | S233 | Uniprot |
| P04049-1 (RAF1) | S259 | Uniprot |
| P04049 (RAF1) | S621 | Uniprot |
| P04083 (ANXA1) | T216 | Uniprot |
| P04626 (ERBB2) | T686 | Uniprot |
| P04637 (TP53) | S378 | Uniprot |
| P04792 (HSPB1) | S78 | Uniprot |
| P04792 (HSPB1) | S82 | Uniprot |
| P04792 (HSPB1) | T143 | Uniprot |
| P05023 (ATP1A1) | S943 | Uniprot |
| P05093 (CYP17A1) | S258 | Uniprot |
| P05114 (HMGN1) | S7 | Uniprot |
| P05114 (HMGN1) | S21 | Uniprot |
| P05114 (HMGN1) | S25 | Uniprot |
| P05204 (HMGN2) | S25 | Uniprot |
| P05204 (HMGN2) | S29 | Uniprot |
| P05549 (TFAP2A) | S239 | Uniprot |
| P05787 (KRT8) | S9 | Uniprot |
| P05787 (KRT8) | S13 | Uniprot |
| P05787 (KRT8) | S24 | Uniprot |
| P05787 (KRT8) | S34 | Uniprot |
| P05787 (KRT8) | S37 | Uniprot |
| P05787 (KRT8) | S43 | Uniprot |
| P05787 (KRT8) | S417 | Uniprot |
| P06239-1 (LCK) | S42 | Uniprot |
| P06241 (FYN) | S21 | Uniprot |
| P06400 (RB1) | S780 | Uniprot |
| P07101-2 (TH) | S36 | Uniprot |
| P07101-1 (TH) | S40 | Uniprot |
| P07101-4 (TH) | S44 | Uniprot |
| P07101-2 (TH) | S67 | Uniprot |
| P07101 (TH) | S71 | Uniprot |
| P07550 (ADRB2) | S345 | Uniprot |
| P07550 (ADRB2) | S346 | Uniprot |
| P07900 (HSP90AA1) | T90 | Uniprot |
| P07949 (RET) | S696 | Uniprot |
| P08034 (GJB1) | S233 | Uniprot |
| P08151 (GLI1) | T374 | Uniprot |
| P08151 (GLI1) | S544 | Uniprot |
| P08151 (GLI1) | S560 | Uniprot |
| P08151 (GLI1) | S640 | Uniprot |
| P08183 (ABCB1) | S667 | Uniprot |
| P08183 (ABCB1) | S671 | Uniprot |
| P08183 (ABCB1) | S683 | Uniprot |
| P08588 (ADRB1) | S312 | Uniprot |
| P08670 (VIM) | S7 | Uniprot |
| P08670 (VIM) | S25 | Uniprot |
| P08670 (VIM) | S39 | Uniprot |
| P08670 (VIM) | S47 | Uniprot |
| P08670 (VIM) | S72 | Uniprot |
| P08670 (VIM) | S73 | Uniprot |
| P08684 (CYP3A4) | S116 | Uniprot |
| P08684 (CYP3A4) | S119 | Uniprot |
| P08684 (CYP3A4) | S134 | Uniprot |
| P08684 (CYP3A4) | S259 | Uniprot |
| P08684 (CYP3A4) | S478 | Uniprot |
| P09038 (FGF2) | T254 | Uniprot |
| P09211 (GSTP1) | S185 | Uniprot |
| P09651 (HNRNPA1) | S199 | Uniprot |
| P09917 (ALOX5) | S272 | Uniprot |
| P09917 (ALOX5) | S524 | Uniprot |
| P0DMV8 (HSPA1A) | S418 | Uniprot |
| P0DN86 (CGB8) | T117 | Uniprot |
| P10071 (GLI3) | S849 | Uniprot |
| P10071 (GLI3) | S865 | Uniprot |
| P10071 (GLI3) | S877 | Uniprot |
| P10071 (GLI3) | S907 | Uniprot |
| P10071 (GLI3) | S980 | Uniprot |
| P10071 (GLI3) | S1006 | Uniprot |
| P10242 (MYB) | S116 | Uniprot |
| P10276 (RARA) | S219 | Uniprot |
| P10276 (RARA) | S369 | Uniprot |
| P10412 (HIST1H1E) | S36 | Uniprot |
| P10636-2 (MAPT) | S137 | Uniprot |
| P10636-6 (MAPT) | S140 | Uniprot |
| P10636-2 (MAPT) | S141 | Uniprot |
| P10636-6 (MAPT) | T147 | Uniprot |
| P10636-6 (MAPT) | T154 | Uniprot |
| P10636-2 (MAPT) | S156 | Uniprot |
| P10636-6 (MAPT) | T173 | Uniprot |
| P10636-8 (MAPT) | S195 | Uniprot |
| P10636-8 (MAPT) | S198 | Uniprot |
| P10636-2 (MAPT) | S204 | Uniprot |
| P10636-8 (MAPT) | T212 | Uniprot |
| P10636-8 (MAPT) | S214 | Uniprot |
| P10636-8 (MAPT) | T245 | Uniprot |
| P10636-8 (MAPT) | S262 | Uniprot |
| P10636-2 (MAPT) | S267 | Uniprot |
| P10636-8 (MAPT) | S293 | Uniprot |
| P10636-6 (MAPT) | S298 | Uniprot |
| P10636-8 (MAPT) | S305 | Uniprot |
| P10636-8 (MAPT) | S316 | Uniprot |
| P10636-2 (MAPT) | S320 | Uniprot |
| P10636-8 (MAPT) | S324 | Uniprot |
| P10636-6 (MAPT) | S351 | Uniprot |
| P10636-8 (MAPT) | S356 | Uniprot |
| P10636-8 (MAPT) | S409 | Uniprot |
| P10636-8 (MAPT) | S416 | Uniprot |
| P10636 (MAPT) | S531 | Uniprot |
| P10636 (MAPT) | T548 | Uniprot |
| P10636 (MAPT) | S579 | Uniprot |
| P10636 (MAPT) | S673 | Uniprot |
| P10636 (MAPT) | S726 | Uniprot |
| P11137-2 (MAP2) | S319 | Uniprot |
| P11137-1 (MAP2) | S1675 | Uniprot |
| P11137 (MAP2) | S1679 | Uniprot |
| P11137 (MAP2) | S1710 | Uniprot |
| P11137 (MAP2) | S1742 | Uniprot |
| P11137 (MAP2) | S1782 | Uniprot |
| P11168-1 (SLC2A2) | S491 | Uniprot |
| P11168-1 (SLC2A2) | S503 | Uniprot |
| P11168-1 (SLC2A2) | S505 | Uniprot |
| P11473 (VDR) | S182 | Uniprot |
| P11717 (IGF2R) | S2347 | Uniprot |
| P11831 (SRF) | T159 | Uniprot |
| P12270 (TPR) | S2108 | Uniprot |
| P12931-1 (SRC) | S17 | Uniprot |
| P12931 (SRC) | Y419 | Uniprot |
| P13224-1 (GP1BB) | S191 | Uniprot |
| P13569 (CFTR) | S660 | Uniprot |
| P13569 (CFTR) | S700 | Uniprot |
| P13569 (CFTR) | S712 | Uniprot |
| P13569-1 (CFTR) | S737 | Uniprot |
| P13569 (CFTR) | S753 | Uniprot |
| P13569 (CFTR) | S768 | Uniprot |
| P13569 (CFTR) | S795 | Uniprot |
| P13569-1 (CFTR) | S813 | Uniprot |
| P13612 (ITGA4) | S1021 | Uniprot |
| P13796 (LCP1) | S5 | Uniprot |
| P13807-1 (GYS1) | S8 | Uniprot |
| P13807-1 (GYS1) | S710 | Uniprot |
| P14136 (GFAP) | T7 | Uniprot |
| P14136 (GFAP) | S8 | Uniprot |
| P14136-1 (GFAP) | S13 | Uniprot |
| P14136-1 (GFAP) | S17 | Uniprot |
| P14136 (GFAP) | S38 | Uniprot |
| P14410 (SI) | S7 | Uniprot |
| P14598 (NCF1) | S320 | Uniprot |
| P14598 (NCF1) | S328 | Uniprot |
| P14598 (NCF1) | S359 | Uniprot |
| P14598 (NCF1) | S370 | Uniprot |
| P14859 (POU2F1) | S385 | Uniprot |
| P15056 (BRAF) | S430 | Uniprot |
| P15311 (EZR) | S66 | Uniprot |
| P15336 (ATF2) | S62 | Uniprot |
| P15924 (DSP) | S2849 | Uniprot |
| P16066 (NPR1) | S529 | Uniprot |
| P16070 (CD44) | S697 | Uniprot |
| P16144 (ITGB4) | S1364 | Uniprot |
| P16220 (CREB1) | S133 | Uniprot |
| P16949 (STMN1) | S16 | Uniprot |
| P16949 (STMN1) | S63 | Uniprot |
| P17302 (GJA1) | S365 | Uniprot |
| P17302 (GJA1) | S368 | Uniprot |
| P17302 (GJA1) | S369 | Uniprot |
| P17302 (GJA1) | S373 | Uniprot |
| P17542 (TAL1) | S122 | Uniprot |
| P17542 (TAL1) | S172 | Uniprot |
| P17600-2 (SYN1) | S9 | Uniprot |
| P17612 (PRKACA) | S11 | Uniprot |
| P17612 (PRKACA) | S15 | Uniprot |
| P17612 (PRKACA) | T49 | Uniprot |
| P17612 (PRKACA) | S54 | Uniprot |
| P17612 (PRKACA) | S140 | Uniprot |
| P17612 (PRKACA) | T198 | Uniprot |
| P17612 (PRKACA) | S213 | Uniprot |
| P17612 (PRKACA) | S260 | Uniprot |
| P17612 (PRKACA) | S326 | Uniprot |
| P17612 (PRKACA) | S339 | Uniprot |
| P17655 (CAPN2) | S369 | Uniprot |
| P17655 (CAPN2) | T370 | Uniprot |
| Substrate | Site | Source |
|---|---|---|
| P08138 (NGFR) | S303 | Uniprot |
| P22694 (PRKACB) | S339 | Uniprot |
| P30518 (AVPR2) | S255 | Uniprot |
| P43699 (NKX2-1) | T9 | Uniprot |
| Q13185 (CBX3) | S93 | Uniprot |
| Q14896 (MYBPC3) | S275 | Uniprot |
| Q14896 (MYBPC3) | S284 | Uniprot |
| Q14896 (MYBPC3) | S304 | Uniprot |
| Q14896 (MYBPC3) | S311 | Uniprot |
| Q92934 (BAD) | S75 | Uniprot |
| Q92934 (BAD) | S99 | Uniprot |
| Q92934 (BAD) | S118 | Uniprot |
Research Backgrounds
Phosphorylates a large number of substrates in the cytoplasm and the nucleus. Regulates the abundance of compartmentalized pools of its regulatory subunits through phosphorylation of PJA2 which binds and ubiquitinates these subunits, leading to their subsequent proteolysis. Phosphorylates CDC25B, ABL1, NFKB1, CLDN3, PSMC5/RPT6, PJA2, RYR2, RORA and VASP. RORA is activated by phosphorylation. Required for glucose-mediated adipogenic differentiation increase and osteogenic differentiation inhibition from osteoblasts. Involved in the regulation of platelets in response to thrombin and collagen; maintains circulating platelets in a resting state by phosphorylating proteins in numerous platelet inhibitory pathways when in complex with NF-kappa-B (NFKB1 and NFKB2) and I-kappa-B-alpha (NFKBIA), but thrombin and collagen disrupt these complexes and free active PRKACA stimulates platelets and leads to platelet aggregation by phosphorylating VASP. Prevents the antiproliferative and anti-invasive effects of alpha-difluoromethylornithine in breast cancer cells when activated. RYR2 channel activity is potentiated by phosphorylation in presence of luminal Ca(2+), leading to reduced amplitude and increased frequency of store overload-induced Ca(2+) release (SOICR) characterized by an increased rate of Ca(2+) release and propagation velocity of spontaneous Ca(2+) waves, despite reduced wave amplitude and resting cytosolic Ca(2+). PSMC5/RPT6 activation by phosphorylation stimulates proteasome. Negatively regulates tight junctions (TJs) in ovarian cancer cells via CLDN3 phosphorylation. NFKB1 phosphorylation promotes NF-kappa-B p50-p50 DNA binding. Involved in embryonic development by down-regulating the Hedgehog (Hh) signaling pathway that determines embryo pattern formation and morphogenesis. Prevents meiosis resumption in prophase-arrested oocytes via CDC25B inactivation by phosphorylation. May also regulate rapid eye movement (REM) sleep in the pedunculopontine tegmental (PPT). Phosphorylates APOBEC3G and AICDA. Isoform 2 phosphorylates and activates ABL1 in sperm flagellum to promote spermatozoa capacitation. Phosphorylates HSF1; this phosphorylation promotes HSF1 nuclear localization and transcriptional activity upon heat shock.
Asn-3 is partially deaminated to Asp giving rise to 2 major isoelectric variants, called CB and CA respectively.
Autophosphorylated. Phosphorylation is enhanced by vitamin K(2). Phosphorylated on threonine and serine residues. Phosphorylation on Thr-198 is required for full activity.
Phosphorylated at Tyr-331 by activated receptor tyrosine kinases EGFR and PDGFR; this increases catalytic efficiency.
Cytoplasm. Cell membrane. Nucleus. Mitochondrion. Membrane>Lipid-anchor.
Note: Translocates into the nucleus (monomeric catalytic subunit). The inactive holoenzyme is found in the cytoplasm. Distributed throughout the cytoplasm in meiotically incompetent oocytes. Associated to mitochondrion as meiotic competence is acquired. Aggregates around the germinal vesicles (GV) at the immature GV stage oocytes (By similarity). Colocalizes with HSF1 in nuclear stress bodies (nSBs) upon heat shock (PubMed:21085490).
Cell projection>Cilium>Flagellum. Cytoplasmic vesicle>Secretory vesicle>Acrosome.
Note: Expressed in the midpiece region of the sperm flagellum (PubMed:10906071). Colocalizes with MROH2B and TCP11 on the acrosome and tail regions in round spermatids and spermatozoa regardless of the capacitation status of the sperm (By similarity).
Isoform 1 is ubiquitous. Isoform 2 is sperm-specific and is enriched in pachytene spermatocytes but is not detected in round spermatids.
A number of inactive tetrameric holoenzymes are produced by the combination of homo- or heterodimers of the different regulatory subunits associated with two catalytic subunits. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits. The cAMP-dependent protein kinase catalytic subunit binds PJA2. Both isoforms 1 and 2 forms activate cAMP-sensitive PKAI and PKAII holoenzymes by interacting with regulatory subunit (R) of PKA, PRKAR1A/PKR1 and PRKAR2A/PKR2, respectively. Interacts with NFKB1, NFKB2 and NFKBIA in platelets; these interactions are disrupted by thrombin and collagen. Binds to ABL1 in spermatozoa and with CDC25B in oocytes. Interacts with APOBEC3G and AICDA. Interacts with RAB13; downstream effector of RAB13 involved in tight junction assembly. Found in a complex at least composed of MROH2B, PRKACA isoform 2 and TCP11 (By similarity). Interacts with MROH2B (By similarity). Isoform 2 interacts with TCP11 (By similarity). Interacts with HSF1.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cAMP subfamily.
Mediates cAMP-dependent signaling triggered by receptor binding to GPCRs. PKA activation regulates diverse cellular processes such as cell proliferation, the cell cycle, differentiation and regulation of microtubule dynamics, chromatin condensation and decondensation, nuclear envelope disassembly and reassembly, as well as regulation of intracellular transport mechanisms and ion flux. Regulates the abundance of compartmentalized pools of its regulatory subunits through phosphorylation of PJA2 which binds and ubiquitinates these subunits, leading to their subsequent proteolysis. Phosphorylates GPKOW which regulates its ability to bind RNA.
Asn-3 is partially deaminated to Asp giving rise to 2 major isoelectric variants, called CB and CA respectively.
Cytoplasm. Cell membrane. Membrane>Lipid-anchor. Nucleus.
Note: Translocates into the nucleus (monomeric catalytic subunit). The inactive holoenzyme is found in the cytoplasm.
Isoform 1 is most abundant in the brain, with low level expression in kidney. Isoform 2 is predominantly expressed in thymus, spleen and kidney. Isoform 3 and isoform 4 are only expressed in the brain.
A number of inactive tetrameric holoenzymes are produced by the combination of homo- or heterodimers of the different regulatory subunits associated with two catalytic subunits. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits (By similarity). The cAMP-dependent protein kinase catalytic subunit binds PJA2. Interacts with GPKOW.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cAMP subfamily.
Phosphorylates a large number of substrates in the cytoplasm and the nucleus.
Testis specific. But important tissues such as brain and ovary have not been analyzed for the content of transcript.
A number of inactive tetrameric holoenzymes are produced by the combination of homo- or heterodimers of the different regulatory subunits associated with two catalytic subunits. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. cAMP subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Gap junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Calcium signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hedgehog signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Neurodegenerative diseases > Parkinson's disease.
· Human Diseases > Neurodegenerative diseases > Prion diseases.
· Human Diseases > Substance dependence > Cocaine addiction.
· Human Diseases > Substance dependence > Amphetamine addiction.
· Human Diseases > Substance dependence > Morphine addiction.
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Infectious diseases: Bacterial > Vibrio cholerae infection.
· Human Diseases > Infectious diseases: Parasitic > Amoebiasis.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cardiovascular diseases > Dilated cardiomyopathy (DCM).
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Circulatory system > Vascular smooth muscle contraction. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
· Organismal Systems > Environmental adaptation > Circadian entrainment.
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Retrograde endocannabinoid signaling. (View pathway)
· Organismal Systems > Nervous system > Glutamatergic synapse.
· Organismal Systems > Nervous system > Cholinergic synapse.
· Organismal Systems > Nervous system > Serotonergic synapse.
· Organismal Systems > Nervous system > GABAergic synapse.
· Organismal Systems > Nervous system > Dopaminergic synapse.
· Organismal Systems > Sensory system > Olfactory transduction.
· Organismal Systems > Sensory system > Taste transduction.
· Organismal Systems > Sensory system > Inflammatory mediator regulation of TRP channels. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Insulin secretion. (View pathway)
· Organismal Systems > Endocrine system > Ovarian steroidogenesis.
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Melanogenesis.
· Organismal Systems > Endocrine system > Thyroid hormone synthesis.
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
· Organismal Systems > Endocrine system > Glucagon signaling pathway.
· Organismal Systems > Endocrine system > Regulation of lipolysis in adipocytes.
· Organismal Systems > Endocrine system > Renin secretion.
· Organismal Systems > Endocrine system > Aldosterone synthesis and secretion.
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
· Organismal Systems > Excretory system > Endocrine and other factor-regulated calcium reabsorption.
· Organismal Systems > Excretory system > Vasopressin-regulated water reabsorption.
· Organismal Systems > Digestive system > Salivary secretion.
· Organismal Systems > Digestive system > Gastric acid secretion.
References
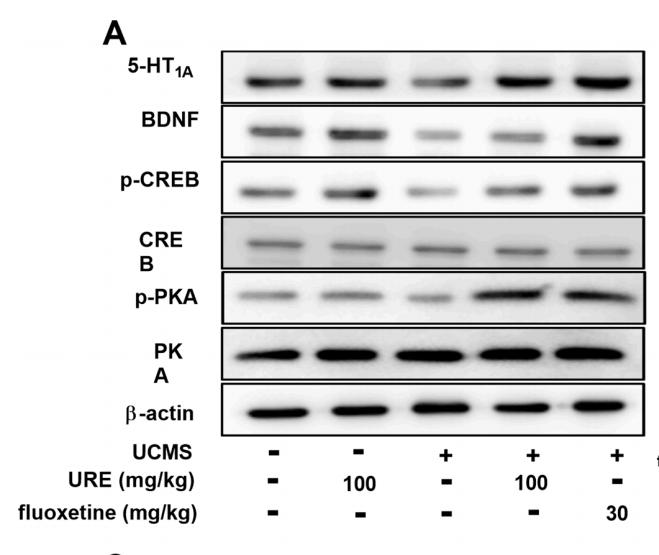
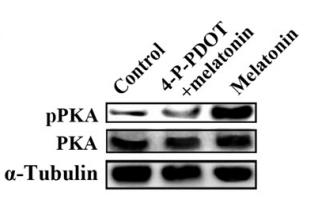
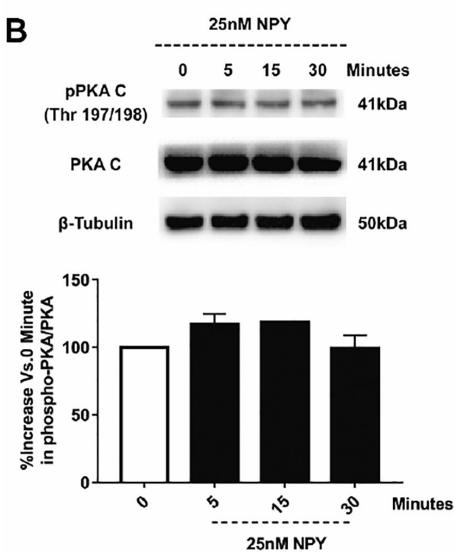
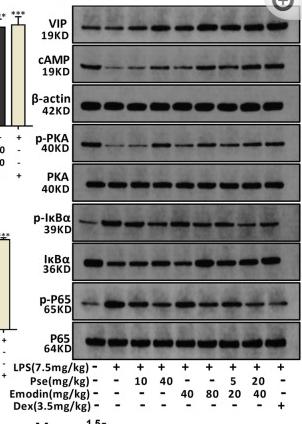
Application: WB Species: mice Sample: hippocampus
Application: WB Species: pig Sample: porcine intramuscular preadipocytes
Application: WB Species: rat Sample: BRL-3A hepatocytes
Application: WB Species: Rat Sample: lung tissues
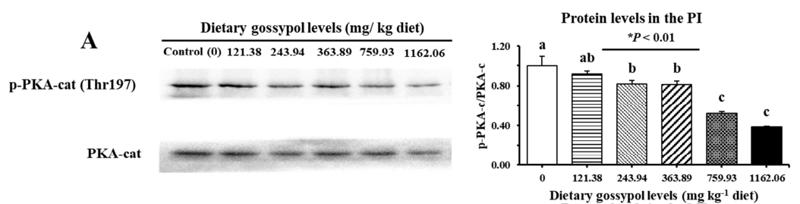
Application: WB Species: fish Sample: proximal intestine
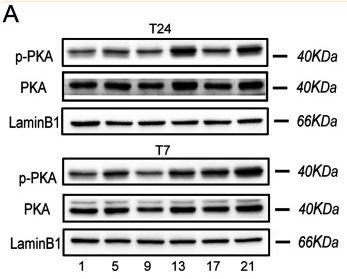
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.