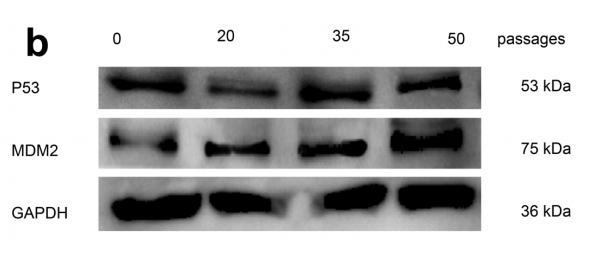
MDM2 Antibody - #AF0208
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0208, RRID:AB_2833395.
Fold/Unfold
ACTFS; Double minute 2 protein; E3 ubiquitin-protein ligase Mdm2; Hdm 2; Hdm2; HDMX; MDM 2; MDM2; MDM2 oncogene E3 ubiquitin protein ligase; Mdm2 p53 E3 ubiquitin protein ligase homolog; Mdm2 transformed 3T3 cell double minute 2 p53 binding protein (mouse) binding protein 104kDa; MDM2_HUMAN; MDM2BP; Mouse Double Minute 2; MTBP; Murine Double Minute Chromosome 2; Oncoprotein Mdm2; p53 Binding Protein Mdm2; p53-binding protein Mdm2; Ubiquitin protein ligase E3 Mdm2; Ubiquitin protein ligase E3 Mdm2;
Immunogens
Ubiquitous. Isoform Mdm2-A, isoform Mdm2-B, isoform Mdm2-C, isoform Mdm2-D, isoform Mdm2-E, isoform Mdm2-F and isoform Mdm2-G are observed in a range of cancers but absent in normal tissues.
- Q00987 MDM2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEVYQVTVYQAGESDTDSFEEDPEISLADYWKCTSCNEMNPPLPSHCNRCWALRENWLPEDKGKDKGEISEKAKLENSTQAEEGFDVPDCKKTIVNDSRESCVEENDDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQDKEESVESSLPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQPIQMIVLTYFP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q00987 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Ubiquitination | Uniprot | ||
| T4 | Phosphorylation | Uniprot | |
| S17 | Phosphorylation | Uniprot | |
| K36 | Ubiquitination | Uniprot | |
| K39 | Ubiquitination | Uniprot | |
| K45 | Ubiquitination | Uniprot | |
| Y60 | Phosphorylation | Uniprot | |
| K70 | Ubiquitination | Uniprot | |
| C77 | S-Nitrosylation | Uniprot | |
| K94 | Ubiquitination | Uniprot | |
| K98 | Ubiquitination | Uniprot | |
| S118 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S121 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| K146 | Ubiquitination | Uniprot | |
| S148 | Phosphorylation | Uniprot | |
| S157 | Phosphorylation | P49137 (MAPKAPK2) | Uniprot |
| T158 | Phosphorylation | Q9NQU5 (PAK6) | Uniprot |
| S160 | Phosphorylation | Uniprot | |
| S166 | Phosphorylation | P48730 (CSNK1D) , P11309 (PIM1) , O00141 (SGK1) , P31749 (AKT1) , P49137 (MAPKAPK2) , P31751 (AKT2) , Q9P1W9 (PIM2) , O43293 (DAPK3) | Uniprot |
| T168 | Phosphorylation | Uniprot | |
| S172 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S176 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| K182 | Acetylation | Uniprot | |
| K185 | Acetylation | Uniprot | |
| S186 | Phosphorylation | Q9NQU5 (PAK6) , Q9P1W9 (PIM2) , P11309 (PIM1) , O43293 (DAPK3) , P31749 (AKT1) , P31751 (AKT2) | Uniprot |
| S188 | Phosphorylation | P31749 (AKT1) | Uniprot |
| S190 | Phosphorylation | Uniprot | |
| S192 | Phosphorylation | Uniprot | |
| S196 | Phosphorylation | Uniprot | |
| T216 | Phosphorylation | Uniprot | |
| T218 | Phosphorylation | Uniprot | |
| S220 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S229 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S240 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S242 | Phosphorylation | Uniprot | |
| S246 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S253 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S256 | Phosphorylation | Uniprot | |
| S260 | Phosphorylation | P68400 (CSNK2A1) , P48730 (CSNK1D) , P53350 (PLK1) | Uniprot |
| S262 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S269 | Phosphorylation | P48730 (CSNK1D) , P68400 (CSNK2A1) | Uniprot |
| Y276 | Phosphorylation | P00519 (ABL1) | Uniprot |
| T279 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| Y281 | Phosphorylation | P12931 (SRC) | Uniprot |
| S286 | Phosphorylation | Uniprot | |
| T288 | Phosphorylation | Uniprot | |
| S290 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| Y302 | Phosphorylation | P12931 (SRC) | Uniprot |
| T306 | Phosphorylation | Uniprot | |
| S307 | Phosphorylation | Uniprot | |
| K334 | Ubiquitination | Uniprot | |
| K336 | Ubiquitination | Uniprot | |
| K338 | Ubiquitination | Uniprot | |
| S342 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| K344 | Ubiquitination | Uniprot | |
| K346 | Ubiquitination | Uniprot | |
| S350 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| T351 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| K363 | Ubiquitination | Uniprot | |
| K364 | Ubiquitination | Uniprot | |
| T365 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S373 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S386 | Phosphorylation | Q13315 (ATM) , P48730 (CSNK1D) | Uniprot |
| S388 | Phosphorylation | Uniprot | |
| Y394 | Phosphorylation | A0A173G4P4 (Abl fusion) , P00519 (ABL1) | Uniprot |
| S395 | Phosphorylation | Q13315 (ATM) | Uniprot |
| Y405 | Phosphorylation | P00519 (ABL1) | Uniprot |
| S406 | Phosphorylation | Uniprot | |
| S407 | Phosphorylation | Q13535 (ATR) | Uniprot |
| T419 | Phosphorylation | P48730 (CSNK1D) | Uniprot |
| S425 | Phosphorylation | Uniprot | |
| S429 | Phosphorylation | Q13315 (ATM) | Uniprot |
| K446 | Sumoylation | Uniprot | |
| K446 | Ubiquitination | Uniprot | |
| K454 | Ubiquitination | Uniprot | |
| K466 | Ubiquitination | Uniprot | |
| K467 | Ubiquitination | Uniprot | |
| K473 | Ubiquitination | Uniprot |
Research Backgrounds
E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as a ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of p53/TP53. Promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation. Component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in stabilizing p53/TP53. Also component of the TRIM28/KAP1-ERBB4-MDM2 complex which links growth factor and DNA damage response pathways. Mediates ubiquitination and subsequent proteasome degradation of DYRK2 in nucleus. Ubiquitinates IGF1R and SNAI1 and promotes them to proteasomal degradation. Ubiquitinates DCX, leading to DCX degradation and reduction of the dendritic spine density of olfactory bulb granule cells (By similarity). Ubiquitinates DLG4, leading to proteasomal degradation of DLG4 which is required for AMPA receptor endocytosis (By similarity).
Phosphorylation on Ser-166 by SGK1 activates ubiquitination of p53/TP53. Phosphorylated at multiple sites near the RING domain by ATM upon DNA damage; this prevents oligomerization and E3 ligase processivity and impedes constitutive p53/TP53 degradation.
Autoubiquitination leads to proteasomal degradation; resulting in p53/TP53 activation it may be regulated by SFN. Also ubiquitinated by TRIM13. Deubiquitinated by USP2 leads to its accumulation and increases deubiquitination and degradation of p53/TP53. Deubiquitinated by USP7 leading to its stabilization.
Nucleus>Nucleoplasm. Cytoplasm. Nucleus>Nucleolus.
Note: Expressed predominantly in the nucleoplasm. Interaction with ARF(P14) results in the localization of both proteins to the nucleolus. The nucleolar localization signals in both ARF(P14) and MDM2 may be necessary to allow efficient nucleolar localization of both proteins. Colocalizes with RASSF1 isoform A in the nucleus.
Ubiquitous. Isoform Mdm2-A, isoform Mdm2-B, isoform Mdm2-C, isoform Mdm2-D, isoform Mdm2-E, isoform Mdm2-F and isoform Mdm2-G are observed in a range of cancers but absent in normal tissues.
Interacts with p53/TP53, TP73/p73, RBL5 and RP11. Binds specifically to RNA. Can interact with RB1, E1A-associated protein EP300 and the E2F1 transcription factor. Forms a ternary complex with p53/TP53 and WWOX. Interacts with CDKN2AIP, RFWD3, USP7, PYHIN1, and RBBP6. Interacts with ARRB1 and ARRB2. Interacts with PSMA3. Found in a trimeric complex with MDM2, MDM4 and USP2. Interacts with USP2 (via N-terminus and C-terminus). Interacts with MDM4. Part of a complex with MDM2, DAXX, RASSF1 and USP7. Part of a complex with DAXX, MDM2 and USP7. Interacts directly with DAXX and USP7. Interacts (via C-terminus) with RASSF1 isoform A (via N-terminus); the interaction is independent of TP53. Interacts with APEX1; leading to its ubiquitination and degradation. Interacts with RYBP; this inhibits ubiquitination of TP53. Identified in a complex with RYBP and p53/TP53. Also component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in regulating p53/TP53 stabilization and activity. Binds directly both p53/TP53 and TRIM28. Component of the TRIM28/KAP1-ERBB4-MDM2 complex involved in connecting growth factor responses with DNA damage. Interacts directly with both TRIM28 and ERBB4 in the complex. Interacts with DYRK2. Interacts with IGF1R. Interacts with TRIM13; the interaction ubiquitinates MDM2 leading to its proteasomal degradation. Interacts with SNAI1; this interaction promotes SNAI1 ubiquitination. Interacts with NOTCH1 (via intracellular domain). Interacts with FHIT. Interacts with RFFL and RNF34; the interaction stabilizes MDM2. Interacts with CDK5RAP3 and CDKN2A/ARF; form a ternary complex involved in regulation of p53/TP53. Interacts with MTA1. Interacts with AARB2. Interacts with MTBP. Interacts with PML. Interacts with TBRG1. Interacts with the 5S RNP which is composed of the 5S RNA, RPL5 and RPL11; the interaction is direct, occurs in the nucleoplasm and negatively regulates MDM2-mediated TP53 ubiquitination and degradation. Interacts with ADGRB1; the interaction results in inhibition of MDM2-mediated ubiquitination and degradation of DLG4/PSD95, promoting DLG4 stability and regulating synaptic plasticity (By similarity). Interacts with RPL23A; this interaction may promote p53/TP53 polyubiquitination.
(Microbial infection) Interacts with herpes virus 8 protein v-IRF4.
(Microbial infection) Interacts with and ubiquitinates HIV-1 Tat.
Region I is sufficient for binding p53 and inhibiting its G1 arrest and apoptosis functions. It also binds p73 and E2F1. Region II contains most of a central acidic region required for interaction with ribosomal protein L5 and a putative C4-type zinc finger. The RING finger domain which coordinates two molecules of zinc interacts specifically with RNA whether or not zinc is present and mediates the heterooligomerization with MDM4. It is also essential for its ubiquitin ligase E3 activity toward p53 and itself.
Belongs to the MDM2/MDM4 family.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Ubiquitin mediated proteolysis. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
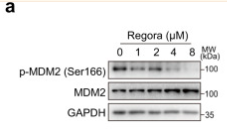
Application: WB Species: Human Sample: HL-7702 cells
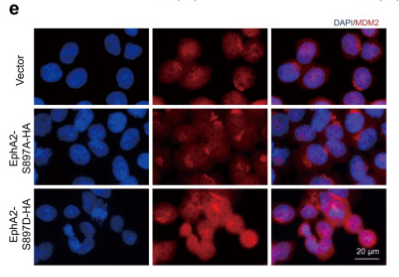
Application: IF/ICC Species: Human Sample: HL-7702 cells
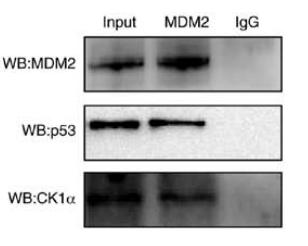
Application: WB Species: Human Sample: MCF-7 cells
Application: WB Species: Mouse Sample:
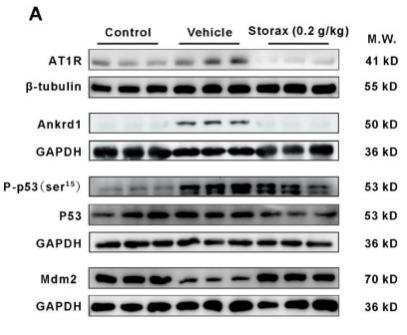
Application: WB Species: Rat Sample: cardiac tissue
Application: WB Species: human Sample: HEL cells.
Application: WB Species: human Sample: HaCaT cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.