Cytokeratin 18 Antibody - #AF0191
| Product: | Cytokeratin 18 Antibody |
| Catalog: | AF0191 |
| Description: | Rabbit polyclonal antibody to Cytokeratin 18 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Sheep, Rabbit |
| Mol.Wt.: | 46kDa; 48kD(Calculated). |
| Uniprot: | P05783 |
| RRID: | AB_2833384 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0191, RRID:AB_2833384.
Fold/Unfold
Cell proliferation inducing gene 46 protein; Cell proliferation inducing protein 46; Cell proliferation-inducing gene 46 protein; CK 18; CK-18; CK18; CYK 18; CYK18; Cytokeratin 18; Cytokeratin endo B; Cytokeratin-18; K 18; K18; K1C18_HUMAN; KA18; Keratin 18; Keratin 18, type I; Keratin D; keratin, type I cytoskeletal 18; Keratin-18; Krt18;
Immunogens
Expressed in colon, placenta, liver and very weakly in exocervix. Increased expression observed in lymph nodes of breast carcinoma.
- P05783 K1C18_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSFTTRSTFSTNYRSLGSVQAPSYGARPVSSAASVYAGAGGSGSRISVSRSTSFRGGMGSGGLATGIAGGLAGMGGIQNEKETMQSLNDRLASYLDRVRSLETENRRLESKIREHLEKKGPQVRDWSHYFKIIEDLRAQIFANTVDNARIVLQIDNARLAADDFRVKYETELAMRQSVENDIHGLRKVIDDTNITRLQLETEIEALKEELLFMKKNHEEEVKGLQAQIASSGLTVEVDAPKSQDLAKIMADIRAQYDELARKNREELDKYWSQQIEESTTVVTTQSAEVGAAETTLTELRRTVQSLEIDLDSMRNLKASLENSLREVEARYALQMEQLNGILLHLESELAQTRAEGQRQAQEYEALLNIKVKLEAEIATYRRLLEDGEDFNLGDALDSSNSMQTIQKTTTRRIVDGKVVSETNDTKVLRH
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P05783 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| M1 | Acetylation | Uniprot | |
| S2 | Acetylation | Uniprot | |
| S2 | Phosphorylation | Uniprot | |
| T4 | Phosphorylation | Uniprot | |
| S7 | Phosphorylation | Uniprot | |
| T8 | Phosphorylation | Uniprot | |
| S10 | Phosphorylation | Uniprot | |
| T11 | Phosphorylation | Uniprot | |
| Y13 | Phosphorylation | Uniprot | |
| S15 | Phosphorylation | Uniprot | |
| S18 | Phosphorylation | Uniprot | |
| S23 | Phosphorylation | Uniprot | |
| Y24 | Phosphorylation | Uniprot | |
| R27 | Methylation | Uniprot | |
| S30 | O-Glycosylation | Uniprot | |
| S30 | Phosphorylation | Uniprot | |
| S31 | O-Glycosylation | Uniprot | |
| S31 | Phosphorylation | Uniprot | |
| S34 | Phosphorylation | P06493 (CDK1) | Uniprot |
| Y36 | Phosphorylation | Uniprot | |
| S42 | Phosphorylation | Uniprot | |
| S44 | Phosphorylation | Uniprot | |
| R45 | Methylation | Uniprot | |
| S47 | Phosphorylation | Uniprot | |
| S49 | O-Glycosylation | Uniprot | |
| S49 | Phosphorylation | Uniprot | |
| R50 | Methylation | Uniprot | |
| S51 | Phosphorylation | Uniprot | |
| T52 | Phosphorylation | Uniprot | |
| S53 | Phosphorylation | P17252 (PRKCA) , Q9UBS0 (RPS6KB2) , P49137 (MAPKAPK2) , Q02156 (PRKCE) , P51812 (RPS6KA3) , Q14012 (CAMK1) , Q9UQM7 (CAMK2A) | Uniprot |
| R55 | Methylation | Uniprot | |
| S60 | Phosphorylation | Uniprot | |
| T65 | Phosphorylation | Uniprot | |
| K81 | Acetylation | Uniprot | |
| K81 | Sumoylation | Uniprot | |
| K81 | Ubiquitination | Uniprot | |
| T83 | Phosphorylation | Uniprot | |
| S86 | Phosphorylation | Uniprot | |
| R90 | Methylation | Uniprot | |
| S93 | Phosphorylation | Uniprot | |
| Y94 | Phosphorylation | Uniprot | |
| R97 | Methylation | Uniprot | |
| S100 | Phosphorylation | Uniprot | |
| S110 | Phosphorylation | Uniprot | |
| K111 | Acetylation | Uniprot | |
| K118 | Methylation | Uniprot | |
| K118 | Ubiquitination | Uniprot | |
| S127 | Phosphorylation | Uniprot | |
| Y129 | Phosphorylation | Uniprot | |
| K131 | Acetylation | Uniprot | |
| K131 | Methylation | Uniprot | |
| K131 | Ubiquitination | Uniprot | |
| K167 | Acetylation | Uniprot | |
| K167 | Methylation | Uniprot | |
| K167 | Sumoylation | Uniprot | |
| K167 | Ubiquitination | Uniprot | |
| Y168 | Phosphorylation | Uniprot | |
| S177 | Phosphorylation | Uniprot | |
| K187 | Acetylation | Uniprot | |
| K187 | Methylation | Uniprot | |
| K187 | Sumoylation | Uniprot | |
| K187 | Ubiquitination | Uniprot | |
| T192 | Phosphorylation | Uniprot | |
| T195 | Phosphorylation | Uniprot | |
| T201 | Phosphorylation | Uniprot | |
| K207 | Sumoylation | Uniprot | |
| K207 | Ubiquitination | Uniprot | |
| K214 | Acetylation | Uniprot | |
| K214 | Ubiquitination | Uniprot | |
| K215 | Ubiquitination | Uniprot | |
| K222 | Acetylation | Uniprot | |
| S230 | Phosphorylation | Uniprot | |
| K241 | Ubiquitination | Uniprot | |
| S242 | Phosphorylation | Uniprot | |
| K247 | Acetylation | Uniprot | |
| K247 | Sumoylation | Uniprot | |
| K247 | Ubiquitination | Uniprot | |
| Y256 | Phosphorylation | Uniprot | |
| K262 | Ubiquitination | Uniprot | |
| K269 | Sumoylation | Uniprot | |
| Y270 | Phosphorylation | Uniprot | |
| S272 | Phosphorylation | Uniprot | |
| T302 | Phosphorylation | Uniprot | |
| S305 | Phosphorylation | Uniprot | |
| S312 | Phosphorylation | Uniprot | |
| K317 | Acetylation | Uniprot | |
| K317 | Sumoylation | Uniprot | |
| K317 | Ubiquitination | Uniprot | |
| S319 | Phosphorylation | Uniprot | |
| S323 | Phosphorylation | Uniprot | |
| Y331 | Phosphorylation | Uniprot | |
| S347 | Phosphorylation | Uniprot | |
| Y363 | Phosphorylation | Uniprot | |
| K370 | Ubiquitination | Uniprot | |
| K372 | Sumoylation | Uniprot | |
| K372 | Ubiquitination | Uniprot | |
| T379 | Phosphorylation | Uniprot | |
| Y380 | Phosphorylation | Uniprot | |
| S398 | Phosphorylation | Uniprot | |
| S399 | Phosphorylation | Uniprot | |
| S401 | Phosphorylation | Uniprot | |
| T404 | Phosphorylation | Uniprot | |
| K407 | Ubiquitination | Uniprot | |
| T409 | Phosphorylation | Uniprot | |
| K417 | Acetylation | Uniprot | |
| K417 | Sumoylation | Uniprot | |
| K417 | Ubiquitination | Uniprot | |
| S420 | Phosphorylation | Uniprot | |
| T422 | Phosphorylation | Uniprot | |
| T425 | Phosphorylation | Uniprot | |
| K426 | Acetylation | Uniprot | |
| K426 | Methylation | Uniprot | |
| K426 | Sumoylation | Uniprot | |
| K426 | Ubiquitination | Uniprot |
Research Backgrounds
Involved in the uptake of thrombin-antithrombin complexes by hepatic cells (By similarity). When phosphorylated, plays a role in filament reorganization. Involved in the delivery of mutated CFTR to the plasma membrane. Together with KRT8, is involved in interleukin-6 (IL-6)-mediated barrier protection.
Phosphorylation at Ser-34 increases during mitosis. Hyperphosphorylated at Ser-53 in diseased cirrhosis liver. Phosphorylation increases by IL-6.
Proteolytically cleaved by caspases during epithelial cell apoptosis. Cleavage occurs at Asp-238 by either caspase-3, caspase-6 or caspase-7.
O-GlcNAcylation increases solubility, and decreases stability by inducing proteasomal degradation.
Cytoplasm>Perinuclear region. Nucleus>Nucleolus.
Expressed in colon, placenta, liver and very weakly in exocervix. Increased expression observed in lymph nodes of breast carcinoma.
Heterotetramer of two type I and two type II keratins. KRT18 associates with KRT8. Interacts with the thrombin-antithrombin complex (By similarity). Interacts with PNN and mutated CFTR. Interacts with YWHAE, YWHAH and YWHAZ only when phosphorylated. Interacts with DNAJB6, TCHP and TRADD. Interacts with FAM83H. Interacts with EPPK1 (By similarity).
(Microbial infection) Interacts with hepatitis C virus/HCV core protein.
Belongs to the intermediate filament family.
Research Fields
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
References
Application: IF/ICC Species: Mouse Sample: ENA-1 cells
Application: WB Species: Rat Sample:
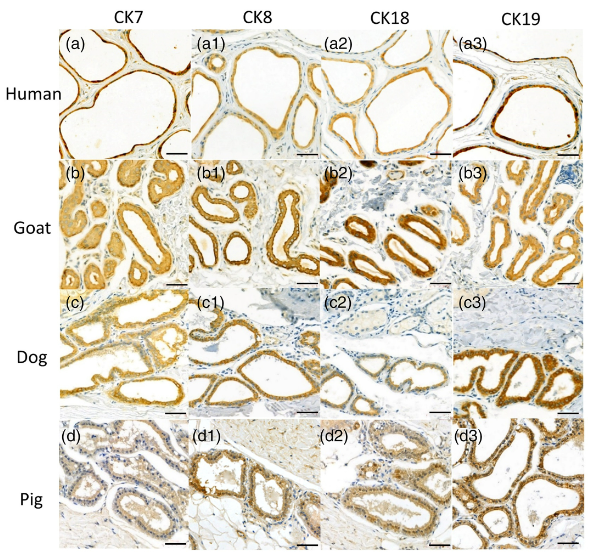
Application: IHC Species: Human,Goat,Dog,Pig Sample: EAC glandular cells
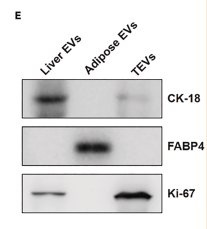
Application: WB Species: Mouse Sample: liver, adipose, and tumor tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.